Explore Mfuzz Clusters
Mfuzz cluster
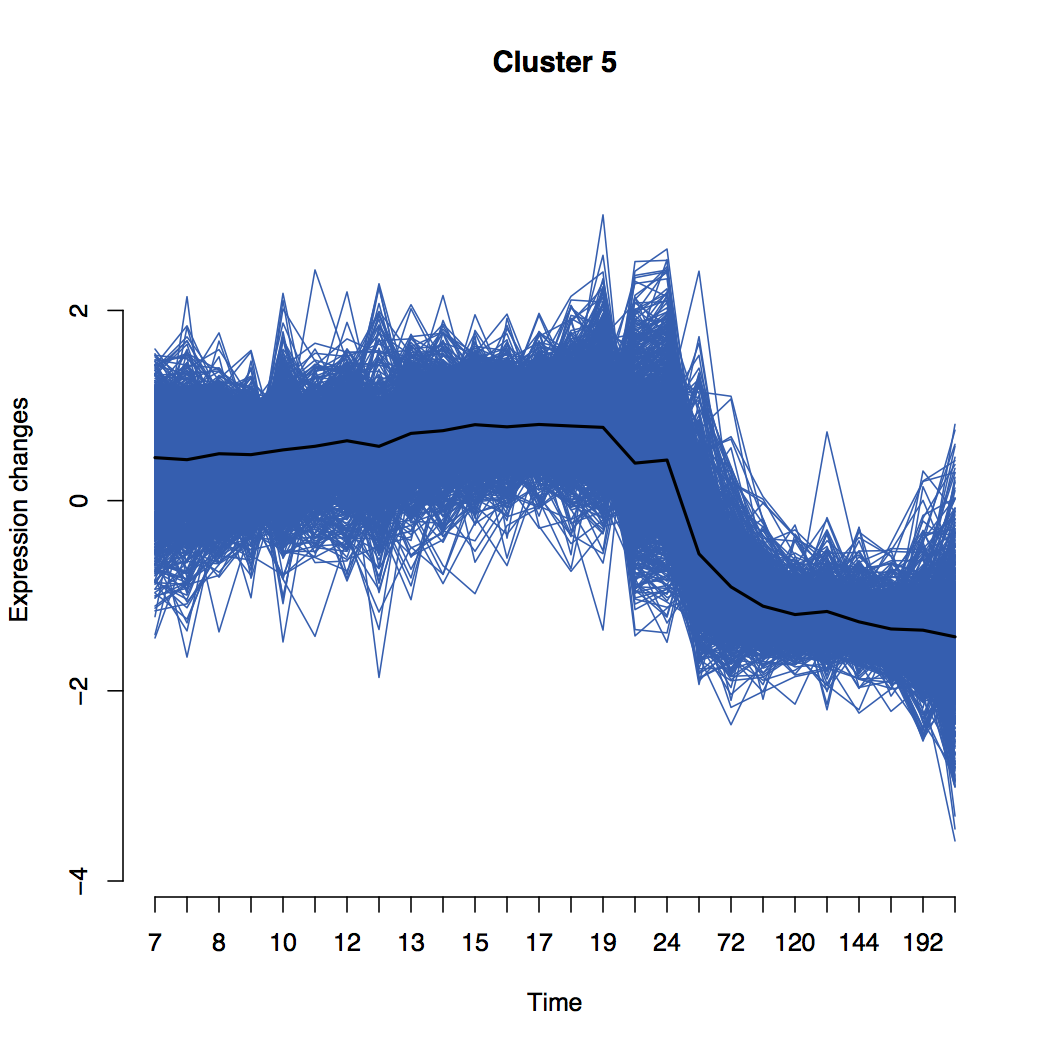
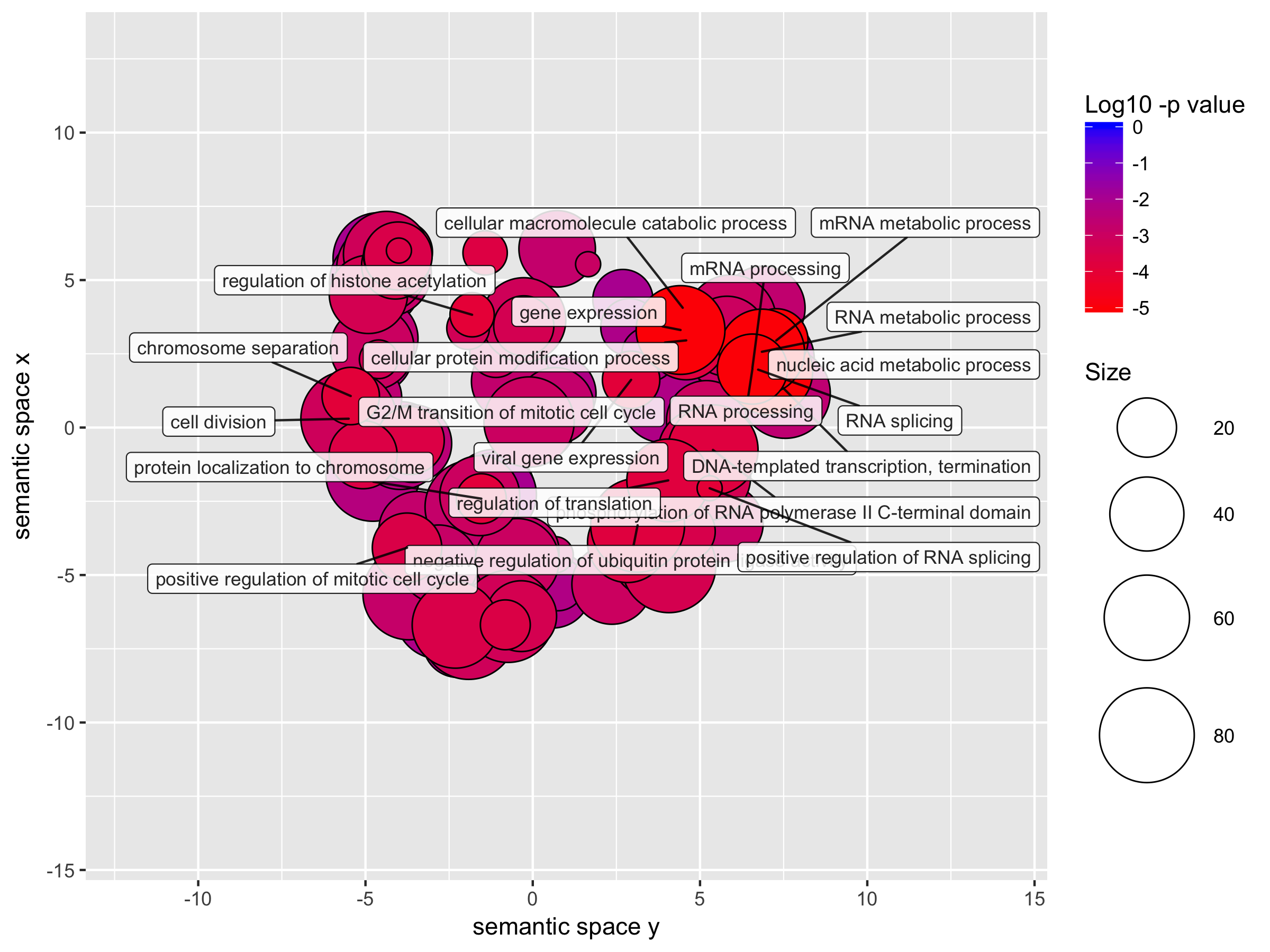
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.220809 | R-6 | 0.998238799649 | E-5 | 1.0 | No_Uniprotmatch | No_Description | gi|156351016|ref|XP_001622324.1|hypothetical protein NEMVEDRAFT_v1g176215 [Nematostella vectensis], 2e-14 |
| other nr_hits | NA | ||||||
| NvERTx.4.222110 | R-1 | 0.812169950334 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.225683 | R-7 | 0.959678339349 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.231825 | R-7 | 0.99904066687 | E-5 | 1.0 | Q6PG31 | RNA-binding protein with serine-rich domain 1 | None |
| other nr_hits | NA | ||||||
| NvERTx.4.233514 | R-8 | 0.838589472182 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.16727 | R-7 | 0.96769390179 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.43539 | R-6 | 0.991969085878 | E-5 | 1.0 | Q54J48 | Probable pyridoxal 5'-phosphate synthase subunit pdx2 | gi|156355414|ref|XP_001623663.1|predicted protein [Nematostella vectensis], 2e-113 |
| other nr_hits | gi|999970198|gb|KXJ09372.1|putative pyridoxal 5'-phosphate synthase subunit PDX2 [Exaiptasia pallida] 6e-72; gi|919079906|ref|XP_013421105.1|PREDICTED: probable pyridoxal 5'-phosphate synthase subunit pdx2 isoform X1 [Lingula anatina] 1e-46; gi|919079908|ref|XP_013421106.1|PREDICTED: probable pyridoxal 5'-phosphate synthase subunit pdx2 isoform X2 [Lingula anatina] 1e-46; gi|919079910|ref|XP_013421107.1|PREDICTED: probable pyridoxal 5'-phosphate synthase subunit pdx2 isoform X3 [Lingula anatina] 1e-46; gi|919079913|ref|XP_013421108.1|PREDICTED: probable pyridoxal 5'-phosphate synthase subunit pdx2 isoform X4 [Lingula anatina] 1e-46; gi|762079701|ref|XP_011413131.1|PREDICTED: uncharacterized protein LOC105317985 [Crassostrea gigas] 3e-45; gi|676455230|ref|XP_009053937.1|hypothetical protein LOTGIDRAFT_116826 [Lottia gigantea] 1e-44; gi|18076243|embl|CAC81976.1|SNO protein [Suberites domuncula] 3e-42; gi|961134681|ref|XP_014787260.1|PREDICTED: probable pyridoxal 5'-phosphate synthase subunit PDX2 [Octopus bimaculoides] 1e-41; | ||||||
| NvERTx.4.63231 | R-7 | 0.951386147581 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.63232 | R-7 | 0.951386147581 | E-5 | 1.0 | Q9JMJ4 | Pre-mRNA-processing factor 19 {ECO:0000305} | gi|156402177|ref|XP_001639467.1|predicted protein [Nematostella vectensis], 2e-261 |
| other nr_hits | gi|999988742|gb|KXJ27812.1|Pre-mRNA-processing factor 19 [Exaiptasia pallida] 7e-224; gi|1005442802|ref|XP_015758740.1|PREDICTED: pre-mRNA-processing factor 19-like [Acropora digitifera] 1e-222; gi|291228334|ref|XP_002734121.1|PREDICTED: pre-mRNA-processing factor 19 [Saccoglossus kowalevskii] 2e-202; gi|957843625|ref|XP_014672184.1|PREDICTED: pre-mRNA-processing factor 19-like isoform X1 [Priapulus caudatus] 5e-199; gi|828195143|ref|XP_012566859.1|PREDICTED: pre-mRNA-processing factor 19-like [Hydra vulgaris] 2e-198; gi|260809518|ref|XP_002599552.1|hypothetical protein BRAFLDRAFT_121774 [Branchiostoma floridae] 5e-196; gi|1126182297|ref|XP_019623659.1|PREDICTED: pre-mRNA-processing factor 19-like [Branchiostoma belcheri] 7e-195; gi|762131114|ref|XP_011448715.1|PREDICTED: pre-mRNA-processing factor 19 [Crassostrea gigas] 2e-192; gi|676447868|ref|XP_009051549.1|hypothetical protein LOTGIDRAFT_103711, partial [Lottia gigantea] 3e-192; | ||||||
| NvERTx.4.63233 | R-7 | 0.951386147581 | E-5 | 1.0 | Q9JMJ4 | Pre-mRNA-processing factor 19 {ECO:0000305} | gi|156402177|ref|XP_001639467.1|predicted protein [Nematostella vectensis], 3e-189 |
| other nr_hits | gi|999988742|gb|KXJ27812.1|Pre-mRNA-processing factor 19 [Exaiptasia pallida] 3e-157; gi|1005442802|ref|XP_015758740.1|PREDICTED: pre-mRNA-processing factor 19-like [Acropora digitifera] 4e-157; gi|780039455|ref|XP_011667864.1|PREDICTED: pre-mRNA-processing factor 19 [Strongylocentrotus purpuratus] 3e-141; gi|291228334|ref|XP_002734121.1|PREDICTED: pre-mRNA-processing factor 19 [Saccoglossus kowalevskii] 3e-141; gi|957843625|ref|XP_014672184.1|PREDICTED: pre-mRNA-processing factor 19-like isoform X1 [Priapulus caudatus] 7e-139; gi|260809518|ref|XP_002599552.1|hypothetical protein BRAFLDRAFT_121774 [Branchiostoma floridae] 1e-138; gi|828195143|ref|XP_012566859.1|PREDICTED: pre-mRNA-processing factor 19-like [Hydra vulgaris] 6e-138; gi|1126182297|ref|XP_019623659.1|PREDICTED: pre-mRNA-processing factor 19-like [Branchiostoma belcheri] 8e-138; gi|734636640|ref|XP_010746969.1|PREDICTED: pre-mRNA-processing factor 19 [Larimichthys crocea] 8e-135; | ||||||
| NvERTx.4.63234 | R-7 | 0.951386147581 | E-5 | 1.0 | Q5ZMA2 | Pre-mRNA-processing factor 19 {ECO:0000305} | gi|156402177|ref|XP_001639467.1|predicted protein [Nematostella vectensis], 1e-76 |
| other nr_hits | gi|1005442802|ref|XP_015758740.1|PREDICTED: pre-mRNA-processing factor 19-like [Acropora digitifera] 4e-67; gi|999988742|gb|KXJ27812.1|Pre-mRNA-processing factor 19 [Exaiptasia pallida] 3e-65; gi|260809518|ref|XP_002599552.1|hypothetical protein BRAFLDRAFT_121774 [Branchiostoma floridae] 4e-64; gi|1126182297|ref|XP_019623659.1|PREDICTED: pre-mRNA-processing factor 19-like [Branchiostoma belcheri] 2e-63; gi|291228334|ref|XP_002734121.1|PREDICTED: pre-mRNA-processing factor 19 [Saccoglossus kowalevskii] 2e-63; gi|82082891|sp|PRP19_CHICK|Q5ZMA2.1|RecName: Full=Pre-mRNA-processing factor 19; AltName: Full=PRP19/PSO4 homolog; AltName: Full=RING-type E3 ubiquitin transferase PRP19 >CAG31141.1 hypothetical protein RCJMB04_2m2 [Gallus gallus] 5e-62; gi|771916210|ref|NP_001034420.2|pre-mRNA-processing factor 19 [Gallus gallus] 5e-62; gi|971396420|ref|XP_015141619.1|PREDICTED: LOW QUALITY PROTEIN: pre-mRNA-processing factor 19-like, partial [Gallus gallus] 1e-61; gi|224050568|ref|XP_002191472.1|PREDICTED: pre-mRNA-processing factor 19 [Taeniopygia guttata] 1e-61; | ||||||
| NvERTx.4.64754 | R-7 | 0.999997599106 | E-5 | 1.0 | Q923G2 | DNA-directed RNA polymerases I, II, and III subunit RPABC3 | gi|156381039|ref|XP_001632074.1|predicted protein [Nematostella vectensis], 1e-79 |
| other nr_hits | gi|999975878|gb|KXJ15052.1|DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Exaiptasia pallida] 2e-75; gi|1005449438|ref|XP_015761937.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Acropora digitifera] 1e-65; gi|556980147|ref|XP_005996821.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Latimeria chalumnae] 6e-65; gi|1041092761|ref|XP_017268875.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Kryptolebias marmoratus] 8e-65; gi|928017255|ref|XP_013855034.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Austrofundulus limnaeus] 8e-65; gi|632968605|ref|XP_007900618.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Callorhinchus milii] 8e-65; gi|1126208297|ref|XP_019637807.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Branchiostoma belcheri] 1e-64; gi|762097053|ref|XP_011431082.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 [Crassostrea gigas] 1e-64; gi|1042251363|ref|XP_017332756.1|PREDICTED: DNA-directed RNA polymerases I, II, and III subunit RPABC3 isoform X1 [Ictalurus punctatus] 1e-64; | ||||||
| NvERTx.4.90311 | R-6 | 0.991969085878 | E-5 | 1.0 | A0A1S3KAG2_LINUN | uncharacterized protein LOC106179969 {ECO:0000313|RefSeq:XP_013419251.1} | gi|1005418391|ref|XP_015780340.1|PREDICTED: uncharacterized protein LOC107357903, partial [Acropora digitifera], 1e-23 |
| other nr_hits | gi|999970188|gb|KXJ09362.1|hypothetical protein AC249_AIPGENE23618 [Exaiptasia pallida] 2e-19; gi|919076339|ref|XP_013419251.1|PREDICTED: uncharacterized protein LOC106179969 [Lingula anatina] 5e-19; gi|676478935|ref|XP_009061583.1|hypothetical protein LOTGIDRAFT_166268 [Lottia gigantea] 4e-18; gi|196003344|ref|XP_002111539.1|hypothetical protein TRIADDRAFT_55664 [Trichoplax adhaerens] 2e-17; gi|1126158406|ref|XP_019614529.1|PREDICTED: uncharacterized protein LOC109462418 [Branchiostoma belcheri] 9e-16; gi|961119648|ref|XP_014782039.1|PREDICTED: uncharacterized protein LOC106877608 [Octopus bimaculoides] 1e-15; gi|918301391|gb|KOF74922.1|hypothetical protein OCBIM_22035487mg [Octopus bimaculoides] 1e-15; gi|1005418637|ref|XP_015762811.1|PREDICTED: uncharacterized protein LOC107341814 [Acropora digitifera] 2e-15; gi|699248764|ref|XP_009860430.1|PREDICTED: uncharacterized protein LOC104266301 [Ciona intestinalis] 1e-14; | ||||||
| NvERTx.4.121669 | R-7 | 0.96769390179 | E-5 | 1.0 | A0A067RN61_ZOONE | Putative ATP-dependent RNA helicase kurz {ECO:0000313|EMBL:KDR21124.1} | gi|156372484|ref|XP_001629067.1|predicted protein [Nematostella vectensis], 1e-63 |
| other nr_hits | gi|999978299|gb|KXJ17473.1|putative ATP-dependent RNA helicase kurz [Exaiptasia pallida] 5e-16; gi|1005454582|ref|XP_015764428.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Acropora digitifera] 4e-10; gi|1072240680|ref|XP_018412785.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Nanorana parkeri] 2e-06; gi|939256655|ref|XP_014248121.1|PREDICTED: probable ATP-dependent RNA helicase kurz [Cimex lectularius] 3e-06; gi|585702769|ref|XP_006823126.1|PREDICTED: probable ATP-dependent RNA helicase DHX37-like [Saccoglossus kowalevskii] 8e-06; gi|646718772|gb|KDR21124.1|putative ATP-dependent RNA helicase kurz [Zootermopsis nevadensis] 1e-05; | ||||||
| NvERTx.4.121674 | R-7 | 0.96769390179 | E-5 | 1.0 | Q8IY37 | Probable ATP-dependent RNA helicase DHX37 | gi|156372484|ref|XP_001629067.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999978299|gb|KXJ17473.1|putative ATP-dependent RNA helicase kurz [Exaiptasia pallida] 0e+00; gi|1005454582|ref|XP_015764428.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Acropora digitifera] 0e+00; gi|657594522|ref|XP_008301878.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Stegastes partitus] 0e+00; gi|1041090730|ref|XP_017267780.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Kryptolebias marmoratus] 0e+00; gi|1129896632|ref|XP_019744561.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Hippocampus comes] 0e+00; gi|1079710450|ref|XP_018529338.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Lates calcarifer] 0e+00; gi|1108947971|ref|XP_019120057.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 isoform X1 [Larimichthys crocea] 0e+00; gi|1108947974|ref|XP_010732547.2|PREDICTED: probable ATP-dependent RNA helicase DHX37 isoform X2 [Larimichthys crocea] 0e+00; gi|698384601|ref|XP_009816357.1|PREDICTED: probable ATP-dependent RNA helicase DHX37, partial [Gavia stellata] 0e+00; | ||||||
| NvERTx.4.121698 | R-7 | 0.96769390179 | E-5 | 1.0 | Q8IY37 | Probable ATP-dependent RNA helicase DHX37 | gi|156372484|ref|XP_001629067.1|predicted protein [Nematostella vectensis], 5e-43 |
| other nr_hits | gi|254553259|ref|NP_001076473.2|probable ATP-dependent RNA helicase DHX37 [Danio rerio] 1e-25; gi|752502583|dbj|BAQ21978.1|Dhx37 protein [Danio rerio] 1e-25; gi|528486753|ref|XP_005172498.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 isoform X1 [Danio rerio] 1e-25; gi|528486755|ref|XP_005172499.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 isoform X2 [Danio rerio] 1e-25; gi|528486757|ref|XP_005172500.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 isoform X3 [Danio rerio] 1e-25; gi|124481892|gb|AAI33161.1|Zgc:158802 protein [Danio rerio] 1e-25; gi|1025163565|ref|XP_016356454.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Sinocyclocheilus anshuiensis] 6e-25; gi|1020503444|ref|XP_016090585.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Sinocyclocheilus grahami] 6e-25; gi|1101597787|ref|XP_018964046.1|PREDICTED: probable ATP-dependent RNA helicase DHX37 [Cyprinus carpio] 1e-24; | ||||||
| NvERTx.4.126354 | R-8 | 0.838589472182 | E-5 | 1.0 | Q8TBZ3 | WD repeat-containing protein 20 | gi|156381922|ref|XP_001632304.1|predicted protein [Nematostella vectensis], 4e-299 |
| other nr_hits | gi|999978330|gb|KXJ17504.1|WD repeat-containing protein 20 [Exaiptasia pallida] 7e-233; gi|1005455111|ref|XP_015764692.1|PREDICTED: WD repeat-containing protein 20-like isoform X1 [Acropora digitifera] 2e-217; gi|1005455113|ref|XP_015764693.1|PREDICTED: WD repeat-containing protein 20-like isoform X2 [Acropora digitifera] 2e-213; gi|632942671|ref|XP_007886536.1|PREDICTED: WD repeat-containing protein 20 [Callorhinchus milii] 1e-162; gi|585637828|ref|XP_006879141.1|PREDICTED: WD repeat-containing protein 20 isoform X3 [Elephantulus edwardii] 6e-162; gi|674105195|ref|XP_008823814.1|PREDICTED: WD repeat-containing protein 20 isoform X2 [Nannospalax galili] 8e-162; gi|674105193|ref|XP_008823813.1|PREDICTED: WD repeat-containing protein 20 isoform X1 [Nannospalax galili] 1e-161; gi|1126208900|ref|XP_019638135.1|PREDICTED: WD repeat-containing protein 20-like isoform X2 [Branchiostoma belcheri] 1e-161; gi|148686714|gb|EDL18661.1|mCG14935, isoform CRA_c, partial [Mus musculus] 1e-161; | ||||||
| NvERTx.4.126355 | R-8 | 0.838589472182 | E-5 | 1.0 | Q8TBZ3 | WD repeat-containing protein 20 | gi|156381922|ref|XP_001632304.1|predicted protein [Nematostella vectensis], 4e-252 |
| other nr_hits | gi|999978330|gb|KXJ17504.1|WD repeat-containing protein 20 [Exaiptasia pallida] 2e-187; gi|1005455111|ref|XP_015764692.1|PREDICTED: WD repeat-containing protein 20-like isoform X1 [Acropora digitifera] 2e-171; gi|1005455113|ref|XP_015764693.1|PREDICTED: WD repeat-containing protein 20-like isoform X2 [Acropora digitifera] 2e-167; gi|585637832|ref|XP_006879143.1|PREDICTED: WD repeat-containing protein 20 isoform X5 [Elephantulus edwardii] 1e-132; gi|674105197|ref|XP_008823815.1|PREDICTED: WD repeat-containing protein 20 isoform X3 [Nannospalax galili] 3e-132; gi|585637834|ref|XP_006879144.1|PREDICTED: WD repeat-containing protein 20 isoform X6 [Elephantulus edwardii] 4e-132; gi|544449557|ref|XP_005562300.1|PREDICTED: WD repeat-containing protein 20 isoform X4 [Macaca fascicularis] 4e-132; gi|602686914|ref|XP_007455686.1|PREDICTED: WD repeat-containing protein 20 isoform X6 [Lipotes vexillifer] 7e-132; gi|1059176737|ref|XP_017744712.1|PREDICTED: WD repeat-containing protein 20 isoform X8 [Rhinopithecus bieti] 1e-131; | ||||||
| NvERTx.4.130008 | R-7 | 0.99904066687 | E-5 | 1.0 | Q28E41 | RNA-binding protein with serine-rich domain 1 | gi|156395189|ref|XP_001636994.1|predicted protein [Nematostella vectensis], 7e-40 |
| other nr_hits | gi|999982572|gb|KXJ21722.1|RNA-binding protein with serine-rich domain 1 [Exaiptasia pallida] 2e-31; gi|1005419074|ref|XP_015750054.1|PREDICTED: RNA-binding protein with serine-rich domain 1-B-like [Acropora digitifera] 3e-26; gi|1126211795|ref|XP_019639707.1|PREDICTED: RNA-binding protein with serine-rich domain 1-like [Branchiostoma belcheri] 2e-23; gi|1049284933|ref|XP_017538976.1|PREDICTED: RNA-binding protein with serine-rich domain 1 isoform X1 [Pygocentrus nattereri] 2e-23; gi|919016038|ref|XP_013392367.1|PREDICTED: RNA-binding protein with serine-rich domain 1-like [Lingula anatina] 3e-23; gi|919021090|ref|XP_013394432.1|PREDICTED: RNA-binding protein with serine-rich domain 1-B-like [Lingula anatina] 3e-23; gi|1002595669|ref|XP_015676790.1|PREDICTED: RNA-binding protein with serine-rich domain 1 [Protobothrops mucrosquamatus] 5e-23; gi|260817443|ref|XP_002603596.1|hypothetical protein BRAFLDRAFT_126915 [Branchiostoma floridae] 5e-23; gi|602634056|ref|XP_007423904.1|PREDICTED: RNA-binding protein with serine-rich domain 1 isoform X1 [Python bivittatus] 5e-23; | ||||||
| NvERTx.4.179266 | R-5 | 0.999989753065 | E-5 | 1.0 | Q9Z1S0 | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta | gi|156404998|ref|XP_001640519.1|predicted protein [Nematostella vectensis], 2e-125 |
| other nr_hits | gi|999986602|gb|KXJ25735.1|Mitotic checkpoint serine/threonine-protein kinase BUB1 beta [Exaiptasia pallida] 4e-97; gi|919080456|ref|XP_013421376.1|PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1-like isoform X1 [Lingula anatina] 9e-83; gi|919018144|ref|XP_013393321.1|PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1-like [Lingula anatina] 1e-82; gi|405972158|gb|EKC36944.1|Mitotic checkpoint serine/threonine-protein kinase BUB1 beta [Crassostrea gigas] 4e-77; gi|762096297|ref|XP_011430673.1|PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1 isoform X2 [Crassostrea gigas] 5e-77; gi|156339952|ref|XP_001620308.1|hypothetical protein NEMVEDRAFT_v1g148546 [Nematostella vectensis] 1e-72; gi|1005433792|ref|XP_015754419.1|PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1 beta-like [Acropora digitifera] 1e-67; gi|1139806060|ref|XP_019923797.1|PREDICTED: mitotic checkpoint serine/threonine-protein kinase BUB1 isoform X1 [Crassostrea gigas] 1e-67; gi|1126216499|ref|XP_019642263.1|PREDICTED: LOW QUALITY PROTEIN: mitotic checkpoint serine/threonine-protein kinase BUB1-like [Branchiostoma belcheri] 3e-66; | ||||||
| NvERTx.4.216672 | R-7 | 0.99904066687 | E-5 | 1.0 | Q5XG24 | RNA-binding protein with serine-rich domain 1-A | gi|156395189|ref|XP_001636994.1|predicted protein [Nematostella vectensis], 7e-31 |
| other nr_hits | gi|999982572|gb|KXJ21722.1|RNA-binding protein with serine-rich domain 1 [Exaiptasia pallida] 5e-25; gi|1005419074|ref|XP_015750054.1|PREDICTED: RNA-binding protein with serine-rich domain 1-B-like [Acropora digitifera] 2e-19; gi|1002595669|ref|XP_015676790.1|PREDICTED: RNA-binding protein with serine-rich domain 1 [Protobothrops mucrosquamatus] 9e-18; gi|927175799|ref|XP_013924314.1|PREDICTED: RNA-binding protein with serine-rich domain 1 [Thamnophis sirtalis] 9e-18; gi|602634056|ref|XP_007423904.1|PREDICTED: RNA-binding protein with serine-rich domain 1 isoform X1 [Python bivittatus] 9e-18; gi|602634062|ref|XP_007423907.1|PREDICTED: RNA-binding protein with serine-rich domain 1 isoform X2 [Python bivittatus] 9e-18; gi|565302662|gb|ETE59100.1|RNA-binding protein with serine-rich domain 1 [Ophiophagus hannah] 9e-18; gi|1126211795|ref|XP_019639707.1|PREDICTED: RNA-binding protein with serine-rich domain 1-like [Branchiostoma belcheri] 1e-17; gi|1121909700|ref|XP_019476131.1|PREDICTED: RNA-binding protein with serine-rich domain 1 isoform X1 [Meleagris gallopavo] 2e-17; | ||||||
| NvERTx.4.57784 | R-7 | 0.999933135756 | E-5 | 1.0 | Q5R8H5 | General transcription factor IIE subunit 1 | gi|156374954|ref|XP_001629848.1|predicted protein [Nematostella vectensis], 2e-197 |
| other nr_hits | gi|999982096|gb|KXJ21248.1|General transcription factor IIE subunit 1 [Exaiptasia pallida] 2e-114; gi|1005440865|ref|XP_015757775.1|PREDICTED: general transcription factor IIE subunit 1-like [Acropora digitifera] 1e-100; gi|828231892|ref|XP_012565299.1|PREDICTED: general transcription factor IIE subunit 1-like [Hydra vulgaris] 5e-81; gi|961067834|ref|XP_014790507.1|PREDICTED: general transcription factor IIE subunit 1-like [Octopus bimaculoides] 2e-54; gi|1061098076|ref|XP_017875724.1|PREDICTED: general transcription factor IIE subunit 1 isoform X1 [Ceratina calcarata] 7e-54; gi|1061098078|ref|XP_017875725.1|PREDICTED: general transcription factor IIE subunit 1 isoform X2 [Ceratina calcarata] 7e-54; gi|1016171706|gb|KZC14742.1|General transcription factor IIE subunit 1 [Dufourea novaeangliae] 7e-54; gi|987922788|ref|XP_015437639.1|PREDICTED: general transcription factor IIE subunit 1 [Dufourea novaeangliae] 9e-54; gi|936684161|ref|XP_014239142.1|PREDICTED: general transcription factor IIE subunit 1 [Trichogramma pretiosum] 9e-54; | ||||||
| NvERTx.4.57785 | R-7 | 0.999933135756 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.57786 | R-7 | 0.999933135756 | E-5 | 1.0 | Q5R8H5 | General transcription factor IIE subunit 1 | gi|156374954|ref|XP_001629848.1|predicted protein [Nematostella vectensis], 2e-197 |
| other nr_hits | gi|999982096|gb|KXJ21248.1|General transcription factor IIE subunit 1 [Exaiptasia pallida] 2e-114; gi|1005440865|ref|XP_015757775.1|PREDICTED: general transcription factor IIE subunit 1-like [Acropora digitifera] 1e-100; gi|828231892|ref|XP_012565299.1|PREDICTED: general transcription factor IIE subunit 1-like [Hydra vulgaris] 5e-81; gi|961067834|ref|XP_014790507.1|PREDICTED: general transcription factor IIE subunit 1-like [Octopus bimaculoides] 2e-54; gi|1061098076|ref|XP_017875724.1|PREDICTED: general transcription factor IIE subunit 1 isoform X1 [Ceratina calcarata] 7e-54; gi|1061098078|ref|XP_017875725.1|PREDICTED: general transcription factor IIE subunit 1 isoform X2 [Ceratina calcarata] 7e-54; gi|1016171706|gb|KZC14742.1|General transcription factor IIE subunit 1 [Dufourea novaeangliae] 7e-54; gi|987922788|ref|XP_015437639.1|PREDICTED: general transcription factor IIE subunit 1 [Dufourea novaeangliae] 9e-54; gi|936684161|ref|XP_014239142.1|PREDICTED: general transcription factor IIE subunit 1 [Trichogramma pretiosum] 9e-54; | ||||||
| NvERTx.4.57787 | R-7 | 0.999933135756 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.64505 | R-6 | 0.833783518064 | E-5 | 1.0 | No_Uniprotmatch | No_Description | gi|156407330|ref|XP_001641497.1|predicted protein [Nematostella vectensis], 2e-45 |
| other nr_hits | NA | ||||||
| NvERTx.4.64506 | R-6 | 0.833783518064 | E-5 | 1.0 | Q9DGD1 | Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C | gi|156407330|ref|XP_001641497.1|predicted protein [Nematostella vectensis], 1e-258 |
| other nr_hits | gi|999988933|gb|KXJ27997.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 4e-136; gi|156361248|ref|XP_001625430.1|predicted protein [Nematostella vectensis] 7e-98; gi|999971871|gb|KXJ11045.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 9e-95; gi|999980069|gb|KXJ19243.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 3e-86; gi|156361954|ref|XP_001625548.1|predicted protein [Nematostella vectensis] 1e-78; gi|828213813|ref|XP_012559455.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A-like, partial [Hydra vulgaris] 8e-78; gi|828205579|ref|XP_004207883.2|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A-like [Hydra vulgaris] 2e-77; gi|1126194748|ref|XP_019630460.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 1e-71; gi|1126194472|ref|XP_019630309.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 2e-71; | ||||||
| NvERTx.4.64507 | R-6 | 0.833783518064 | E-5 | 1.0 | Q9DGD1 | Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C | gi|156407330|ref|XP_001641497.1|predicted protein [Nematostella vectensis], 1e-247 |
| other nr_hits | gi|999988933|gb|KXJ27997.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 3e-136; gi|156361248|ref|XP_001625430.1|predicted protein [Nematostella vectensis] 1e-97; gi|999971871|gb|KXJ11045.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 8e-95; gi|999980069|gb|KXJ19243.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 2e-86; gi|156361954|ref|XP_001625548.1|predicted protein [Nematostella vectensis] 1e-78; gi|828213813|ref|XP_012559455.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A-like, partial [Hydra vulgaris] 7e-78; gi|828205579|ref|XP_004207883.2|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A-like [Hydra vulgaris] 2e-77; gi|1126194748|ref|XP_019630460.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 1e-71; gi|1126194472|ref|XP_019630309.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 2e-71; | ||||||
| NvERTx.4.64508 | R-6 | 0.833783518064 | E-5 | 1.0 | Q9DGD1 | Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C | gi|156407330|ref|XP_001641497.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999988933|gb|KXJ27997.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 4e-148; gi|156361248|ref|XP_001625430.1|predicted protein [Nematostella vectensis] 9e-98; gi|999971871|gb|KXJ11045.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 1e-94; gi|999980069|gb|KXJ19243.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 3e-86; gi|156361954|ref|XP_001625548.1|predicted protein [Nematostella vectensis] 2e-78; gi|828213813|ref|XP_012559455.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A-like, partial [Hydra vulgaris] 1e-77; gi|828205579|ref|XP_004207883.2|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A-like [Hydra vulgaris] 2e-77; gi|1126194748|ref|XP_019630460.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 1e-71; gi|1126194472|ref|XP_019630309.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 3e-71; | ||||||
| NvERTx.4.65210 | R-7 | 0.999972700165 | E-5 | 1.0 | Q7T3C6 | Cleavage and polyadenylation specificity factor subunit 5 | gi|999982921|gb|KXJ22067.1|Cleavage and polyadenylation specificity factor subunit 5 [Exaiptasia pallida], 8e-110 |
| other nr_hits | gi|156401665|ref|XP_001639411.1|predicted protein [Nematostella vectensis] 6e-102; gi|928015859|ref|XP_013883401.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Austrofundulus limnaeus] 1e-93; gi|1041066486|ref|XP_017296387.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Kryptolebias marmoratus] 2e-93; gi|1126192420|ref|XP_019629205.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 isoform X2 [Branchiostoma belcheri] 3e-93; gi|551512488|ref|XP_005807759.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Xiphophorus maculatus] 5e-93; gi|47230369|embl|CAF99562.1|unnamed protein product, partial [Tetraodon nigroviridis] 6e-93; gi|410913111|ref|XP_003970032.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Takifugu rubripes] 6e-93; gi|1007735724|ref|XP_015808521.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Nothobranchius furzeri] 8e-93; gi|638265547|ref|NP_001279900.1|cleavage and polyadenylation specificity factor subunit 5 [Callorhinchus milii] 8e-93; | ||||||
| NvERTx.4.65211 | R-7 | 0.999972700165 | E-5 | 1.0 | Q7T3C6 | Cleavage and polyadenylation specificity factor subunit 5 | gi|999982921|gb|KXJ22067.1|Cleavage and polyadenylation specificity factor subunit 5 [Exaiptasia pallida], 5e-53 |
| other nr_hits | gi|928015859|ref|XP_013883401.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Austrofundulus limnaeus] 5e-44; gi|1041066486|ref|XP_017296387.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Kryptolebias marmoratus] 9e-44; gi|156401665|ref|XP_001639411.1|predicted protein [Nematostella vectensis] 1e-43; gi|551512488|ref|XP_005807759.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Xiphophorus maculatus] 2e-43; gi|47230369|embl|CAF99562.1|unnamed protein product, partial [Tetraodon nigroviridis] 3e-43; gi|929129335|ref|XP_013982434.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5-like [Salmo salar] 3e-43; gi|410913111|ref|XP_003970032.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Takifugu rubripes] 3e-43; gi|1007735724|ref|XP_015808521.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5 [Nothobranchius furzeri] 3e-43; gi|929307389|ref|XP_014031272.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 5-like [Salmo salar] 3e-43; | ||||||
| NvERTx.4.93480 | R-6 | 0.993870616354 | E-5 | 1.0 | Q6DGP2 | Pre-mRNA-splicing factor syf2 | gi|156376452|ref|XP_001630374.1|predicted protein [Nematostella vectensis], 8e-45 |
| other nr_hits | gi|919016657|ref|XP_013392701.1|PREDICTED: pre-mRNA-splicing factor SYF2-like [Lingula anatina] 1e-31; gi|919044556|ref|XP_013405461.1|PREDICTED: pre-mRNA-splicing factor SYF2-like [Lingula anatina] 1e-31; gi|72078047|ref|XP_788894.1|PREDICTED: pre-mRNA-splicing factor SYF2 isoform X1 [Strongylocentrotus purpuratus] 3e-28; gi|780183433|ref|XP_011662337.1|PREDICTED: pre-mRNA-splicing factor SYF2 isoform X2 [Strongylocentrotus purpuratus] 3e-28; gi|343459117|gb|AEM37717.1|pre-mRNA-splicing factor, partial [Epinephelus bruneus] 9e-28; gi|556954182|ref|XP_005988809.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Latimeria chalumnae] 1e-27; gi|1129948870|ref|XP_019733872.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Hippocampus comes] 2e-27; gi|586463644|ref|XP_006862457.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Chrysochloris asiatica] 2e-27; gi|554823369|ref|XP_005922364.1|PREDICTED: pre-mRNA-splicing factor SYF2 isoform X1 [Haplochromis burtoni] 2e-27; | ||||||
| NvERTx.4.93483 | R-6 | 0.993870616354 | E-5 | 1.0 | Q6DGP2 | Pre-mRNA-splicing factor syf2 | gi|156376452|ref|XP_001630374.1|predicted protein [Nematostella vectensis], 1e-97 |
| other nr_hits | gi|919016657|ref|XP_013392701.1|PREDICTED: pre-mRNA-splicing factor SYF2-like [Lingula anatina] 3e-62; gi|919044556|ref|XP_013405461.1|PREDICTED: pre-mRNA-splicing factor SYF2-like [Lingula anatina] 3e-62; gi|974099783|ref|XP_015248555.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Cyprinodon variegatus] 1e-61; gi|658916123|ref|XP_008397822.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Poecilia reticulata] 2e-61; gi|551528137|ref|XP_005815538.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Xiphophorus maculatus] 2e-61; gi|1025423281|ref|XP_016517353.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Poecilia formosa] 3e-61; gi|343459117|gb|AEM37717.1|pre-mRNA-splicing factor, partial [Epinephelus bruneus] 4e-61; gi|1129948870|ref|XP_019733872.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Hippocampus comes] 5e-61; gi|961849377|ref|XP_014889515.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Poecilia latipinna] 1e-60; | ||||||
| NvERTx.4.93485 | R-6 | 0.993870616354 | E-5 | 1.0 | Q8AVQ6 | Pre-mRNA-splicing factor syf2 | gi|156376452|ref|XP_001630374.1|predicted protein [Nematostella vectensis], 2e-24 |
| other nr_hits | gi|919016657|ref|XP_013392701.1|PREDICTED: pre-mRNA-splicing factor SYF2-like [Lingula anatina] 2e-16; gi|919044556|ref|XP_013405461.1|PREDICTED: pre-mRNA-splicing factor SYF2-like [Lingula anatina] 2e-16; gi|443711197|gb|ELU05061.1|hypothetical protein CAPTEDRAFT_20848 [Capitella teleta] 1e-15; gi|730369325|gb|KHJ41169.1|SYF2 splicing factor [Trichuris suis] 1e-14; gi|669305276|gb|KFD48826.1|hypothetical protein M513_10310, partial [Trichuris suis] 1e-14; gi|556954182|ref|XP_005988809.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Latimeria chalumnae] 2e-14; gi|1121796873|ref|XP_019505667.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Hipposideros armiger] 3e-14; gi|669226523|embl|CDW52180.1|pre mrna splicing factor syf2 [Trichuris trichiura] 3e-14; gi|725584710|ref|XP_010343735.1|PREDICTED: pre-mRNA-splicing factor SYF2 [Saimiri boliviensis boliviensis] 3e-14; | ||||||
| NvERTx.4.182373 | R-6 | 0.689016419705 | E-5 | 1.0 | Q7ZXH9 | Alpha-endosulfine | gi|156358403|ref|XP_001624509.1|predicted protein [Nematostella vectensis], 2e-47 |
| other nr_hits | gi|999970941|gb|KXJ10115.1|Alpha-endosulfine [Exaiptasia pallida] 2e-25; gi|1005423873|ref|XP_015773650.1|PREDICTED: alpha-endosulfine-like [Acropora digitifera] 1e-19; gi|1143363801|ref|XP_019935127.1|PREDICTED: cAMP-regulated phosphoprotein 19-like [Paralichthys olivaceus] 1e-12; gi|961999511|ref|XP_014875930.1|PREDICTED: cAMP-regulated phosphoprotein 19-like [Poecilia latipinna] 2e-12; gi|1035270922|ref|XP_016888240.1|PREDICTED: cAMP-regulated phosphoprotein 19 isoform X1 [Cynoglossus semilaevis] 3e-12; gi|928040533|ref|XP_013869023.1|PREDICTED: cAMP-regulated phosphoprotein 19 [Austrofundulus limnaeus] 3e-12; gi|657750592|ref|XP_008310712.1|PREDICTED: cAMP-regulated phosphoprotein 19 isoform X2 [Cynoglossus semilaevis] 3e-12; gi|1072260260|ref|XP_018415406.1|PREDICTED: alpha-endosulfine [Nanorana parkeri] 4e-12; gi|961859158|ref|XP_014845094.1|PREDICTED: cAMP-regulated phosphoprotein 19-like [Poecilia mexicana] 4e-12; | ||||||
| NvERTx.4.182374 | R-6 | 0.689016419705 | E-5 | 1.0 | Q7ZXH9 | Alpha-endosulfine | gi|156358403|ref|XP_001624509.1|predicted protein [Nematostella vectensis], 4e-44 |
| other nr_hits | gi|999970941|gb|KXJ10115.1|Alpha-endosulfine [Exaiptasia pallida] 1e-25; gi|1005423873|ref|XP_015773650.1|PREDICTED: alpha-endosulfine-like [Acropora digitifera] 7e-20; gi|961999511|ref|XP_014875930.1|PREDICTED: cAMP-regulated phosphoprotein 19-like [Poecilia latipinna] 2e-13; gi|928040533|ref|XP_013869023.1|PREDICTED: cAMP-regulated phosphoprotein 19 [Austrofundulus limnaeus] 3e-13; gi|1143363801|ref|XP_019935127.1|PREDICTED: cAMP-regulated phosphoprotein 19-like [Paralichthys olivaceus] 3e-13; gi|961859158|ref|XP_014845094.1|PREDICTED: cAMP-regulated phosphoprotein 19-like [Poecilia mexicana] 3e-13; gi|632938265|ref|XP_007904364.1|PREDICTED: cAMP-regulated phosphoprotein 19 [Callorhinchus milii] 5e-13; gi|638256737|ref|NP_001279837.1|cAMP-regulated phosphoprotein 19 [Callorhinchus milii] 5e-13; gi|1072260260|ref|XP_018415406.1|PREDICTED: alpha-endosulfine [Nanorana parkeri] 6e-13; | ||||||
| NvERTx.4.182379 | R-6 | 0.689016419705 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.220470 | R-6 | 0.833783518064 | E-5 | 1.0 | O77836 | Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A | gi|156407330|ref|XP_001641497.1|predicted protein [Nematostella vectensis], 3e-51 |
| other nr_hits | gi|999988933|gb|KXJ27997.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 4e-36; gi|1126194472|ref|XP_019630309.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 3e-25; gi|1126194470|ref|XP_019630308.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like, partial [Branchiostoma belcheri] 4e-25; gi|1126194474|ref|XP_019630310.1|PREDICTED: LOW QUALITY PROTEIN: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 4e-25; gi|828208616|ref|XP_012557703.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A-like [Hydra vulgaris] 5e-25; gi|1126194748|ref|XP_019630460.1|PREDICTED: alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase C-like [Branchiostoma belcheri] 1e-24; gi|156361954|ref|XP_001625548.1|predicted protein [Nematostella vectensis] 2e-24; gi|156361248|ref|XP_001625430.1|predicted protein [Nematostella vectensis] 4e-24; gi|999971871|gb|KXJ11045.1|Alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A [Exaiptasia pallida] 5e-24; | ||||||
| NvERTx.4.96133 | R-5 | 0.760022494489 | E-5 | 1.0 | Q5PQP2 | Receptor-binding cancer antigen expressed on SiSo cells | gi|156364762|ref|XP_001626514.1|predicted protein [Nematostella vectensis], 3e-50 |
| other nr_hits | gi|999977604|gb|KXJ16778.1|Receptor-binding cancer antigen expressed on SiSo cells [Exaiptasia pallida] 3e-24; gi|1005487508|ref|XP_015780345.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells-like [Acropora digitifera] 1e-20; gi|999967026|gb|KXJ06200.1|Receptor-binding cancer antigen expressed on SiSo cells, partial [Exaiptasia pallida] 1e-13; gi|260785905|ref|XP_002588000.1|hypothetical protein BRAFLDRAFT_125399 [Branchiostoma floridae] 2e-11; gi|1126215484|ref|XP_019641708.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells-like [Branchiostoma belcheri] 9e-11; gi|831551654|ref|XP_012728426.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Fundulus heteroclitus] 2e-09; gi|974100660|ref|XP_015249039.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Cyprinodon variegatus] 5e-09; gi|1007727215|ref|XP_015804867.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Nothobranchius furzeri] 8e-09; gi|1129983978|ref|XP_019748648.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Hippocampus comes] 2e-08; | ||||||
| NvERTx.4.96142 | R-5 | 0.760022494489 | E-5 | 1.0 | Q5PQP2 | Receptor-binding cancer antigen expressed on SiSo cells | gi|156364762|ref|XP_001626514.1|predicted protein [Nematostella vectensis], 3e-50 |
| other nr_hits | gi|999977604|gb|KXJ16778.1|Receptor-binding cancer antigen expressed on SiSo cells [Exaiptasia pallida] 3e-24; gi|1005487508|ref|XP_015780345.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells-like [Acropora digitifera] 1e-20; gi|999967026|gb|KXJ06200.1|Receptor-binding cancer antigen expressed on SiSo cells, partial [Exaiptasia pallida] 1e-13; gi|260785905|ref|XP_002588000.1|hypothetical protein BRAFLDRAFT_125399 [Branchiostoma floridae] 2e-11; gi|1126215484|ref|XP_019641708.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells-like [Branchiostoma belcheri] 9e-11; gi|831551654|ref|XP_012728426.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Fundulus heteroclitus] 2e-09; gi|974100660|ref|XP_015249039.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Cyprinodon variegatus] 5e-09; gi|1007727215|ref|XP_015804867.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Nothobranchius furzeri] 8e-09; gi|1129983978|ref|XP_019748648.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Hippocampus comes] 2e-08; | ||||||
| NvERTx.4.96143 | R-5 | 0.760022494489 | E-5 | 1.0 | Q5PQP2 | Receptor-binding cancer antigen expressed on SiSo cells | gi|156364762|ref|XP_001626514.1|predicted protein [Nematostella vectensis], 3e-50 |
| other nr_hits | gi|999977604|gb|KXJ16778.1|Receptor-binding cancer antigen expressed on SiSo cells [Exaiptasia pallida] 3e-24; gi|1005487508|ref|XP_015780345.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells-like [Acropora digitifera] 1e-20; gi|999967026|gb|KXJ06200.1|Receptor-binding cancer antigen expressed on SiSo cells, partial [Exaiptasia pallida] 1e-13; gi|260785905|ref|XP_002588000.1|hypothetical protein BRAFLDRAFT_125399 [Branchiostoma floridae] 2e-11; gi|1126215484|ref|XP_019641708.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells-like [Branchiostoma belcheri] 9e-11; gi|831551654|ref|XP_012728426.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Fundulus heteroclitus] 2e-09; gi|974100660|ref|XP_015249039.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Cyprinodon variegatus] 5e-09; gi|1007727215|ref|XP_015804867.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Nothobranchius furzeri] 8e-09; gi|1129983978|ref|XP_019748648.1|PREDICTED: receptor-binding cancer antigen expressed on SiSo cells [Hippocampus comes] 2e-08; | ||||||
| NvERTx.4.106026 | R-5 | 0.974955284452 | E-5 | 1.0 | Q96K80 | Zinc finger CCCH domain-containing protein 10 | gi|156384276|ref|XP_001633257.1|predicted protein [Nematostella vectensis], 1e-95 |
| other nr_hits | gi|999978809|gb|KXJ17983.1|Zinc finger CCCH domain-containing protein 10 [Exaiptasia pallida] 1e-56; gi|1005446779|ref|XP_015760679.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like [Acropora digitifera] 2e-51; gi|221102276|ref|XP_002167360.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like [Hydra vulgaris] 2e-29; gi|524891418|ref|XP_005102242.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X1 [Aplysia californica] 1e-26; gi|524891420|ref|XP_005102243.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X2 [Aplysia californica] 1e-26; gi|260783958|ref|XP_002587037.1|hypothetical protein BRAFLDRAFT_80617 [Branchiostoma floridae] 1e-25; gi|908488962|ref|XP_013094685.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X1 [Biomphalaria glabrata] 4e-25; gi|908488964|ref|XP_013094686.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X2 [Biomphalaria glabrata] 4e-25; gi|676585887|gb|KFO74723.1|Zinc finger CCCH domain-containing protein 10, partial [Cuculus canorus] 2e-24; | ||||||
| NvERTx.4.106027 | R-5 | 0.974955284452 | E-5 | 1.0 | Q96K80 | Zinc finger CCCH domain-containing protein 10 | gi|156384276|ref|XP_001633257.1|predicted protein [Nematostella vectensis], 1e-95 |
| other nr_hits | gi|999978809|gb|KXJ17983.1|Zinc finger CCCH domain-containing protein 10 [Exaiptasia pallida] 1e-56; gi|1005446779|ref|XP_015760679.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like [Acropora digitifera] 2e-51; gi|221102276|ref|XP_002167360.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like [Hydra vulgaris] 2e-29; gi|524891418|ref|XP_005102242.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X1 [Aplysia californica] 1e-26; gi|524891420|ref|XP_005102243.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X2 [Aplysia californica] 1e-26; gi|260783958|ref|XP_002587037.1|hypothetical protein BRAFLDRAFT_80617 [Branchiostoma floridae] 1e-25; gi|908488962|ref|XP_013094685.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X1 [Biomphalaria glabrata] 4e-25; gi|908488964|ref|XP_013094686.1|PREDICTED: zinc finger CCCH domain-containing protein 10-like isoform X2 [Biomphalaria glabrata] 4e-25; gi|676585887|gb|KFO74723.1|Zinc finger CCCH domain-containing protein 10, partial [Cuculus canorus] 2e-24; | ||||||
| NvERTx.4.114874 | R-5 | 0.965766464089 | E-5 | 1.0 | Q7T012 | Haloacid dehalogenase-like hydrolase domain-containing protein 3 | gi|156400164|ref|XP_001638870.1|predicted protein [Nematostella vectensis], 6e-118 |
| other nr_hits | gi|1126201683|ref|XP_019634196.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Branchiostoma belcheri] 2e-35; gi|1005458274|ref|XP_015766268.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Acropora digitifera] 4e-35; gi|260822308|ref|XP_002606544.1|hypothetical protein BRAFLDRAFT_131071 [Branchiostoma floridae] 8e-35; gi|260793048|ref|XP_002591525.1|hypothetical protein BRAFLDRAFT_131048 [Branchiostoma floridae] 2e-34; gi|390349654|ref|XP_003727254.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Strongylocentrotus purpuratus] 8e-33; gi|554834127|ref|XP_005927586.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 isoform X1 [Haplochromis burtoni] 1e-32; gi|554834129|ref|XP_005927587.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 isoform X2 [Haplochromis burtoni] 1e-32; gi|919074991|ref|XP_013418637.1|PREDICTED: uncharacterized protein LOC106179520 [Lingula anatina] 3e-32; gi|498976240|ref|XP_004548820.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 isoform X1 [Maylandia zebra] 3e-32; | ||||||
| NvERTx.4.114875 | R-5 | 0.965766464089 | E-5 | 1.0 | Q7T012 | Haloacid dehalogenase-like hydrolase domain-containing protein 3 | gi|156400164|ref|XP_001638870.1|predicted protein [Nematostella vectensis], 6e-118 |
| other nr_hits | gi|1126201683|ref|XP_019634196.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Branchiostoma belcheri] 2e-35; gi|1005458274|ref|XP_015766268.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Acropora digitifera] 4e-35; gi|260822308|ref|XP_002606544.1|hypothetical protein BRAFLDRAFT_131071 [Branchiostoma floridae] 8e-35; gi|260793048|ref|XP_002591525.1|hypothetical protein BRAFLDRAFT_131048 [Branchiostoma floridae] 2e-34; gi|390349654|ref|XP_003727254.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Strongylocentrotus purpuratus] 8e-33; gi|554834127|ref|XP_005927586.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 isoform X1 [Haplochromis burtoni] 1e-32; gi|554834129|ref|XP_005927587.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 isoform X2 [Haplochromis burtoni] 1e-32; gi|919074991|ref|XP_013418637.1|PREDICTED: uncharacterized protein LOC106179520 [Lingula anatina] 3e-32; gi|498976240|ref|XP_004548820.1|PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 isoform X1 [Maylandia zebra] 3e-32; | ||||||
| NvERTx.4.80536 | R-7 | 0.999803152319 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.80537 | R-7 | 0.999803152319 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.80538 | R-7 | 0.999803152319 | E-5 | 1.0 | Q9I8J6 | Probable rRNA-processing protein EBP2 | gi|156400824|ref|XP_001638992.1|predicted protein [Nematostella vectensis], 1e-105 |
| other nr_hits | gi|999987568|gb|KXJ26681.1|putative rRNA-processing protein EBP2 [Exaiptasia pallida] 6e-62; gi|1005425721|ref|XP_015748313.1|PREDICTED: probable rRNA-processing protein EBP2 [Acropora digitifera] 7e-47; gi|926631852|ref|XP_013781670.1|PREDICTED: probable rRNA-processing protein EBP2 [Limulus polyphemus] 1e-45; gi|919054946|ref|XP_013409419.1|PREDICTED: probable rRNA-processing protein EBP2 [Lingula anatina] 1e-45; gi|919086381|ref|XP_013380332.1|PREDICTED: probable rRNA-processing protein EBP2 isoform X1 [Lingula anatina] 2e-45; gi|1139792874|ref|XP_011428759.2|PREDICTED: probable rRNA-processing protein EBP2 [Crassostrea gigas] 1e-43; gi|291234480|ref|XP_002737177.1|PREDICTED: probable rRNA-processing protein EBP2-like [Saccoglossus kowalevskii] 4e-43; gi|62857943|ref|NP_001017151.1|EBNA1 binding protein 2 [Xenopus tropicalis] 1e-42; gi|1101636311|ref|XP_018978361.1|PREDICTED: probable rRNA-processing protein EBP2 [Cyprinus carpio] 3e-42; | ||||||
| NvERTx.4.80539 | R-7 | 0.999803152319 | E-5 | 1.0 | Q9I8J6 | Probable rRNA-processing protein EBP2 | gi|156400824|ref|XP_001638992.1|predicted protein [Nematostella vectensis], 1e-74 |
| other nr_hits | gi|999987568|gb|KXJ26681.1|putative rRNA-processing protein EBP2 [Exaiptasia pallida] 1e-40; gi|1005425721|ref|XP_015748313.1|PREDICTED: probable rRNA-processing protein EBP2 [Acropora digitifera] 4e-35; gi|926631852|ref|XP_013781670.1|PREDICTED: probable rRNA-processing protein EBP2 [Limulus polyphemus] 2e-32; gi|62857943|ref|NP_001017151.1|EBNA1 binding protein 2 [Xenopus tropicalis] 1e-31; gi|919086381|ref|XP_013380332.1|PREDICTED: probable rRNA-processing protein EBP2 isoform X1 [Lingula anatina] 1e-31; gi|919054946|ref|XP_013409419.1|PREDICTED: probable rRNA-processing protein EBP2 [Lingula anatina] 1e-31; gi|1101636311|ref|XP_018978361.1|PREDICTED: probable rRNA-processing protein EBP2 [Cyprinus carpio] 2e-31; gi|1069344666|ref|XP_018111868.1|PREDICTED: probable rRNA-processing protein EBP2 isoform X1 [Xenopus laevis] 6e-31; gi|1050382685|gb|OCT84880.1|hypothetical protein XELAEV_18023040mg [Xenopus laevis] 6e-31; | ||||||
| NvERTx.4.80540 | R-7 | 0.999803152319 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
