Explore Mfuzz Clusters
Mfuzz cluster
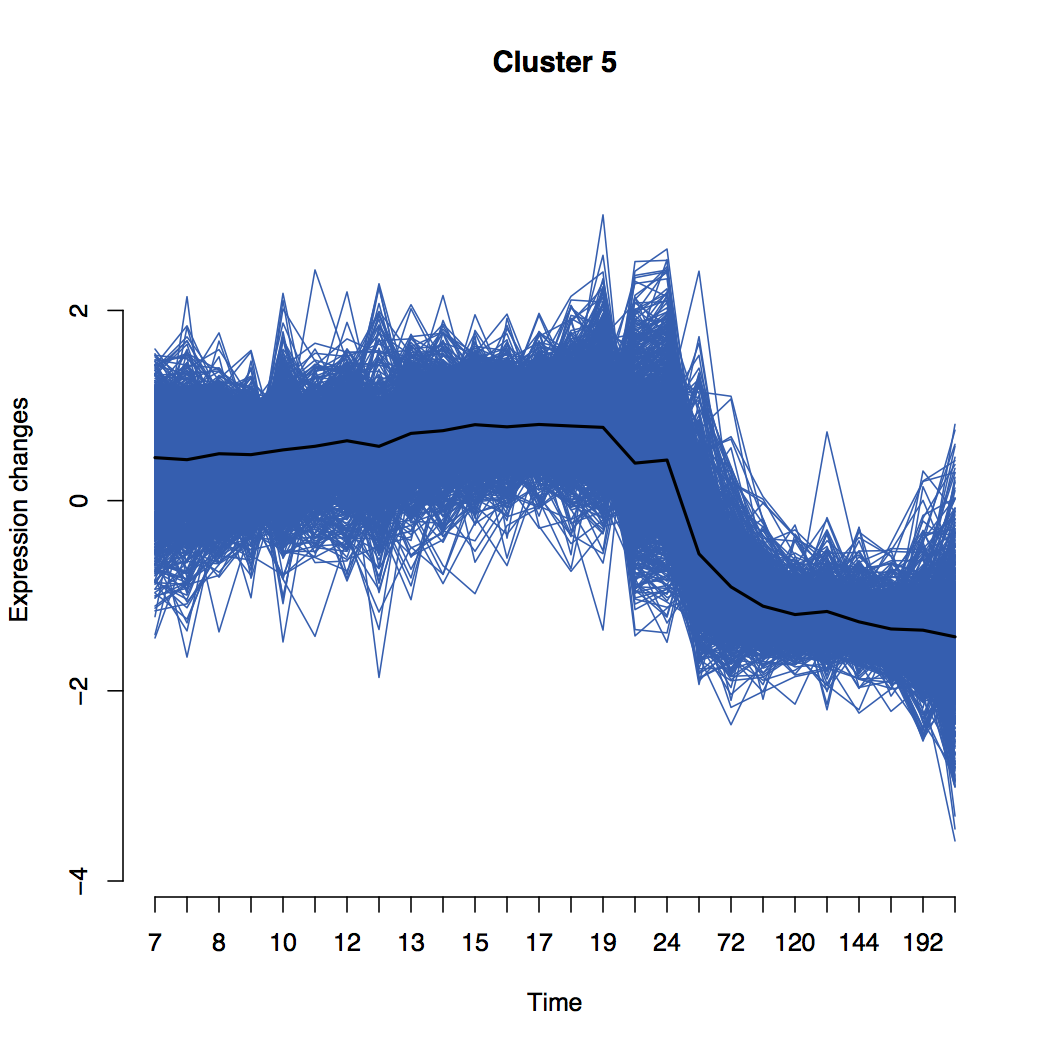
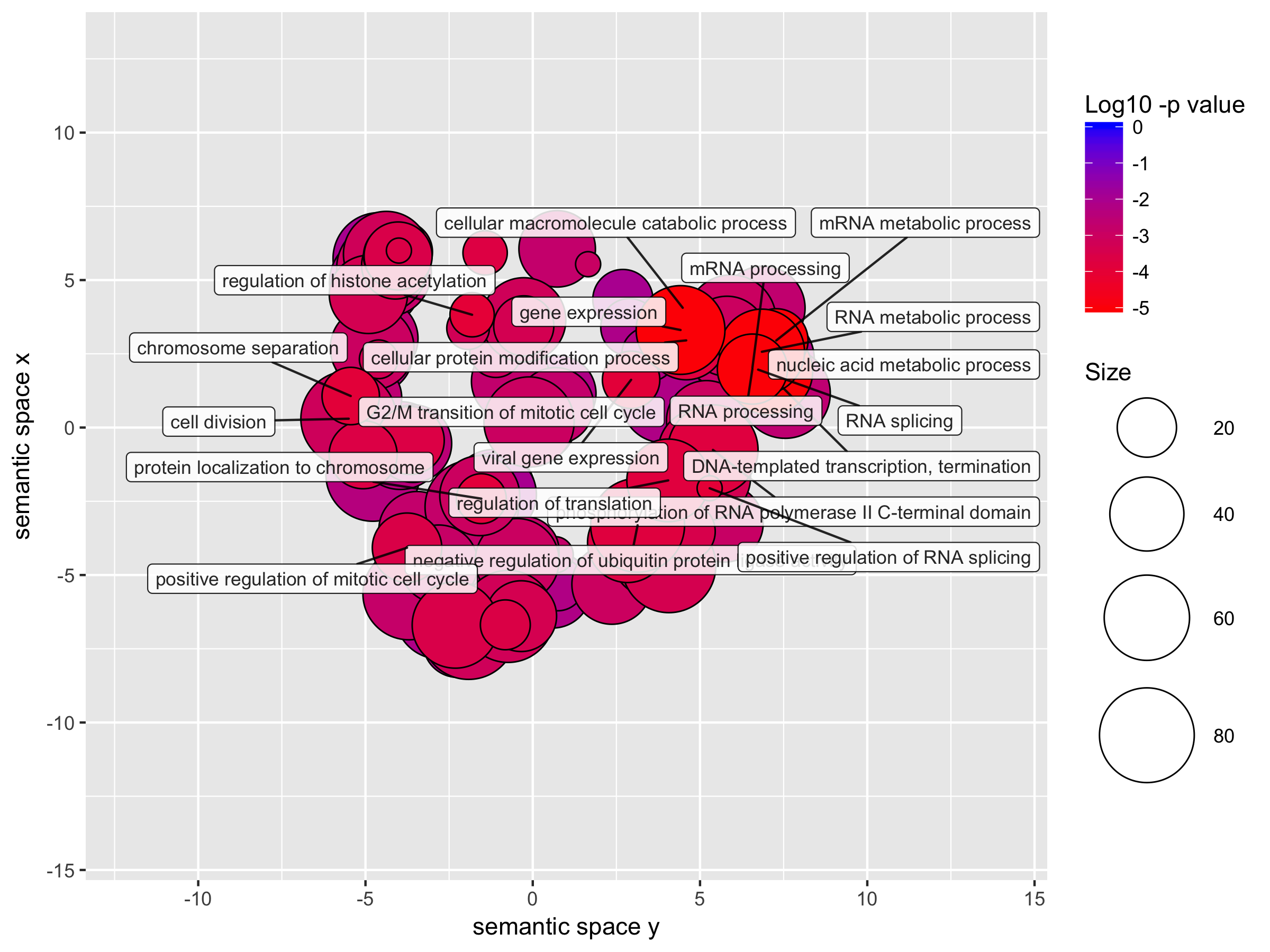
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.100683 | R-7 | 0.726819351321 | E-5 | 1.0 | Q9NPI0 | Transmembrane protein 138 | gi|156351265|ref|XP_001622434.1|predicted protein [Nematostella vectensis], 8e-75 |
| other nr_hits | gi|999974283|gb|KXJ13457.1|Transmembrane protein 138 [Exaiptasia pallida] 4e-69; gi|1005468116|ref|XP_015770972.1|PREDICTED: transmembrane protein 138-like [Acropora digitifera] 4e-62; gi|260837171|ref|XP_002613579.1|hypothetical protein BRAFLDRAFT_277366 [Branchiostoma floridae] 2e-53; gi|1126173512|ref|XP_019618891.1|PREDICTED: transmembrane protein 138-like isoform X1 [Branchiostoma belcheri] 4e-53; gi|919058957|ref|XP_013411319.1|PREDICTED: transmembrane protein 138-like [Lingula anatina] 6e-49; gi|443734898|gb|ELU18754.1|hypothetical protein CAPTEDRAFT_147200 [Capitella teleta] 4e-48; gi|390360669|ref|XP_001185328.2|PREDICTED: transmembrane protein 138 [Strongylocentrotus purpuratus] 5e-48; gi|504181758|ref|XP_004599374.1|PREDICTED: transmembrane protein 138 [Ochotona princeps] 8e-46; gi|345310983|ref|XP_001517871.2|PREDICTED: transmembrane protein 138 isoform X1 [Ornithorhynchus anatinus] 8e-46; | ||||||
| NvERTx.4.100692 | R-7 | 0.726819351321 | E-5 | 1.0 | Q9NPI0 | Transmembrane protein 138 | gi|156351265|ref|XP_001622434.1|predicted protein [Nematostella vectensis], 8e-75 |
| other nr_hits | gi|999974283|gb|KXJ13457.1|Transmembrane protein 138 [Exaiptasia pallida] 4e-69; gi|1005468116|ref|XP_015770972.1|PREDICTED: transmembrane protein 138-like [Acropora digitifera] 4e-62; gi|260837171|ref|XP_002613579.1|hypothetical protein BRAFLDRAFT_277366 [Branchiostoma floridae] 2e-53; gi|1126173512|ref|XP_019618891.1|PREDICTED: transmembrane protein 138-like isoform X1 [Branchiostoma belcheri] 4e-53; gi|919058957|ref|XP_013411319.1|PREDICTED: transmembrane protein 138-like [Lingula anatina] 6e-49; gi|443734898|gb|ELU18754.1|hypothetical protein CAPTEDRAFT_147200 [Capitella teleta] 4e-48; gi|390360669|ref|XP_001185328.2|PREDICTED: transmembrane protein 138 [Strongylocentrotus purpuratus] 5e-48; gi|504181758|ref|XP_004599374.1|PREDICTED: transmembrane protein 138 [Ochotona princeps] 8e-46; gi|345310983|ref|XP_001517871.2|PREDICTED: transmembrane protein 138 isoform X1 [Ornithorhynchus anatinus] 8e-46; | ||||||
| NvERTx.4.100684 | R-7 | 0.726819351321 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.100685 | R-7 | 0.726819351321 | E-5 | 1.0 | Q9NPI0 | Transmembrane protein 138 | gi|156351265|ref|XP_001622434.1|predicted protein [Nematostella vectensis], 8e-75 |
| other nr_hits | gi|999974283|gb|KXJ13457.1|Transmembrane protein 138 [Exaiptasia pallida] 4e-69; gi|1005468116|ref|XP_015770972.1|PREDICTED: transmembrane protein 138-like [Acropora digitifera] 4e-62; gi|260837171|ref|XP_002613579.1|hypothetical protein BRAFLDRAFT_277366 [Branchiostoma floridae] 2e-53; gi|1126173512|ref|XP_019618891.1|PREDICTED: transmembrane protein 138-like isoform X1 [Branchiostoma belcheri] 4e-53; gi|919058957|ref|XP_013411319.1|PREDICTED: transmembrane protein 138-like [Lingula anatina] 6e-49; gi|443734898|gb|ELU18754.1|hypothetical protein CAPTEDRAFT_147200 [Capitella teleta] 4e-48; gi|390360669|ref|XP_001185328.2|PREDICTED: transmembrane protein 138 [Strongylocentrotus purpuratus] 5e-48; gi|504181758|ref|XP_004599374.1|PREDICTED: transmembrane protein 138 [Ochotona princeps] 8e-46; gi|345310983|ref|XP_001517871.2|PREDICTED: transmembrane protein 138 isoform X1 [Ornithorhynchus anatinus] 8e-46; | ||||||
| NvERTx.4.100686 | R-7 | 0.726819351321 | E-5 | 1.0 | Q9NPI0 | Transmembrane protein 138 | gi|156351265|ref|XP_001622434.1|predicted protein [Nematostella vectensis], 8e-75 |
| other nr_hits | gi|999974283|gb|KXJ13457.1|Transmembrane protein 138 [Exaiptasia pallida] 4e-69; gi|1005468116|ref|XP_015770972.1|PREDICTED: transmembrane protein 138-like [Acropora digitifera] 4e-62; gi|260837171|ref|XP_002613579.1|hypothetical protein BRAFLDRAFT_277366 [Branchiostoma floridae] 2e-53; gi|1126173512|ref|XP_019618891.1|PREDICTED: transmembrane protein 138-like isoform X1 [Branchiostoma belcheri] 4e-53; gi|919058957|ref|XP_013411319.1|PREDICTED: transmembrane protein 138-like [Lingula anatina] 6e-49; gi|443734898|gb|ELU18754.1|hypothetical protein CAPTEDRAFT_147200 [Capitella teleta] 4e-48; gi|390360669|ref|XP_001185328.2|PREDICTED: transmembrane protein 138 [Strongylocentrotus purpuratus] 5e-48; gi|504181758|ref|XP_004599374.1|PREDICTED: transmembrane protein 138 [Ochotona princeps] 8e-46; gi|345310983|ref|XP_001517871.2|PREDICTED: transmembrane protein 138 isoform X1 [Ornithorhynchus anatinus] 8e-46; | ||||||
| NvERTx.4.100687 | R-7 | 0.726819351321 | E-5 | 1.0 | Q9NPI0 | Transmembrane protein 138 | gi|156351265|ref|XP_001622434.1|predicted protein [Nematostella vectensis], 8e-75 |
| other nr_hits | gi|999974283|gb|KXJ13457.1|Transmembrane protein 138 [Exaiptasia pallida] 4e-69; gi|1005468116|ref|XP_015770972.1|PREDICTED: transmembrane protein 138-like [Acropora digitifera] 4e-62; gi|260837171|ref|XP_002613579.1|hypothetical protein BRAFLDRAFT_277366 [Branchiostoma floridae] 2e-53; gi|1126173512|ref|XP_019618891.1|PREDICTED: transmembrane protein 138-like isoform X1 [Branchiostoma belcheri] 4e-53; gi|919058957|ref|XP_013411319.1|PREDICTED: transmembrane protein 138-like [Lingula anatina] 6e-49; gi|443734898|gb|ELU18754.1|hypothetical protein CAPTEDRAFT_147200 [Capitella teleta] 4e-48; gi|390360669|ref|XP_001185328.2|PREDICTED: transmembrane protein 138 [Strongylocentrotus purpuratus] 5e-48; gi|504181758|ref|XP_004599374.1|PREDICTED: transmembrane protein 138 [Ochotona princeps] 8e-46; gi|345310983|ref|XP_001517871.2|PREDICTED: transmembrane protein 138 isoform X1 [Ornithorhynchus anatinus] 8e-46; | ||||||
| NvERTx.4.100688 | R-7 | 0.726819351321 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.100689 | R-7 | 0.726819351321 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.100690 | R-7 | 0.726819351321 | E-5 | 1.0 | Q9NPI0 | Transmembrane protein 138 | gi|156351265|ref|XP_001622434.1|predicted protein [Nematostella vectensis], 8e-75 |
| other nr_hits | gi|999974283|gb|KXJ13457.1|Transmembrane protein 138 [Exaiptasia pallida] 4e-69; gi|1005468116|ref|XP_015770972.1|PREDICTED: transmembrane protein 138-like [Acropora digitifera] 4e-62; gi|260837171|ref|XP_002613579.1|hypothetical protein BRAFLDRAFT_277366 [Branchiostoma floridae] 2e-53; gi|1126173512|ref|XP_019618891.1|PREDICTED: transmembrane protein 138-like isoform X1 [Branchiostoma belcheri] 4e-53; gi|919058957|ref|XP_013411319.1|PREDICTED: transmembrane protein 138-like [Lingula anatina] 6e-49; gi|443734898|gb|ELU18754.1|hypothetical protein CAPTEDRAFT_147200 [Capitella teleta] 4e-48; gi|390360669|ref|XP_001185328.2|PREDICTED: transmembrane protein 138 [Strongylocentrotus purpuratus] 5e-48; gi|504181758|ref|XP_004599374.1|PREDICTED: transmembrane protein 138 [Ochotona princeps] 8e-46; gi|345310983|ref|XP_001517871.2|PREDICTED: transmembrane protein 138 isoform X1 [Ornithorhynchus anatinus] 8e-46; | ||||||
| NvERTx.4.100691 | R-7 | 0.726819351321 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.102155 | R-6 | 0.926161571225 | E-5 | 1.0 | D3ZAA9 | MAGUK p55 subfamily member 2 | gi|156374008|ref|XP_001629601.1|predicted protein [Nematostella vectensis], 9e-289 |
| other nr_hits | gi|999986377|gb|KXJ25516.1|MAGUK p55 subfamily member 6 [Exaiptasia pallida] 2e-238; gi|1126188276|ref|XP_019626945.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X21 [Branchiostoma belcheri] 2e-175; gi|1126188278|ref|XP_019626946.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X22 [Branchiostoma belcheri] 3e-174; gi|1126188274|ref|XP_019626944.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X20 [Branchiostoma belcheri] 2e-171; gi|1126188272|ref|XP_019626943.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X19 [Branchiostoma belcheri] 3e-171; gi|1126188270|ref|XP_019626942.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X18 [Branchiostoma belcheri] 1e-168; gi|557016585|ref|XP_006008722.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X1 [Latimeria chalumnae] 1e-162; gi|557016588|ref|XP_006008723.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X2 [Latimeria chalumnae] 8e-162; gi|530624900|ref|XP_005301999.1|PREDICTED: MAGUK p55 subfamily member 2 isoform X1 [Chrysemys picta bellii] 4e-160; | ||||||
| NvERTx.4.102156 | R-6 | 0.926161571225 | E-5 | 1.0 | Q14168 | MAGUK p55 subfamily member 2 | gi|156374008|ref|XP_001629601.1|predicted protein [Nematostella vectensis], 3e-27 |
| other nr_hits | gi|1126188254|ref|XP_019626933.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X11 [Branchiostoma belcheri] 3e-21; gi|1126188236|ref|XP_019626924.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X2 [Branchiostoma belcheri] 3e-21; gi|1126188252|ref|XP_019626932.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X10 [Branchiostoma belcheri] 3e-21; gi|1126188270|ref|XP_019626942.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X18 [Branchiostoma belcheri] 3e-21; gi|1126188264|ref|XP_019626939.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X15 [Branchiostoma belcheri] 3e-21; gi|1126188238|ref|XP_019626925.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X3 [Branchiostoma belcheri] 3e-21; gi|1126188258|ref|XP_019626935.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X12 [Branchiostoma belcheri] 3e-21; gi|1126188250|ref|XP_019626931.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X9 [Branchiostoma belcheri] 3e-21; gi|1126188260|ref|XP_019626937.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X13 [Branchiostoma belcheri] 3e-21; | ||||||
| NvERTx.4.102157 | R-6 | 0.926161571225 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.102158 | R-6 | 0.926161571225 | E-5 | 1.0 | D3ZAA9 | MAGUK p55 subfamily member 2 | gi|156374008|ref|XP_001629601.1|predicted protein [Nematostella vectensis], 1e-289 |
| other nr_hits | gi|999986377|gb|KXJ25516.1|MAGUK p55 subfamily member 6 [Exaiptasia pallida] 2e-238; gi|1126188276|ref|XP_019626945.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X21 [Branchiostoma belcheri] 2e-176; gi|1126188278|ref|XP_019626946.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X22 [Branchiostoma belcheri] 2e-175; gi|1126188274|ref|XP_019626944.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X20 [Branchiostoma belcheri] 1e-172; gi|1126188272|ref|XP_019626943.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X19 [Branchiostoma belcheri] 2e-172; gi|1126188270|ref|XP_019626942.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X18 [Branchiostoma belcheri] 8e-170; gi|557016585|ref|XP_006008722.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X1 [Latimeria chalumnae] 1e-162; gi|557016588|ref|XP_006008723.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X2 [Latimeria chalumnae] 1e-162; gi|530624900|ref|XP_005301999.1|PREDICTED: MAGUK p55 subfamily member 2 isoform X1 [Chrysemys picta bellii] 7e-161; | ||||||
| NvERTx.4.102159 | R-6 | 0.926161571225 | E-5 | 1.0 | Q9JLB0 | MAGUK p55 subfamily member 6 | gi|156374008|ref|XP_001629601.1|predicted protein [Nematostella vectensis], 9e-209 |
| other nr_hits | gi|999986377|gb|KXJ25516.1|MAGUK p55 subfamily member 6 [Exaiptasia pallida] 9e-164; gi|1126188276|ref|XP_019626945.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X21 [Branchiostoma belcheri] 3e-135; gi|1126188278|ref|XP_019626946.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X22 [Branchiostoma belcheri] 4e-134; gi|1126188274|ref|XP_019626944.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X20 [Branchiostoma belcheri] 4e-134; gi|1126188272|ref|XP_019626943.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X19 [Branchiostoma belcheri] 1e-133; gi|1126188270|ref|XP_019626942.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X18 [Branchiostoma belcheri] 3e-131; gi|113674066|ref|NP_001038242.1|MAGUK p55 subfamily member 6 [Danio rerio] 1e-125; gi|528509761|ref|XP_005170070.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X1 [Danio rerio] 1e-125; gi|557016585|ref|XP_006008722.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X1 [Latimeria chalumnae] 3e-125; | ||||||
| NvERTx.4.102160 | R-6 | 0.926161571225 | E-5 | 1.0 | D3ZAA9 | MAGUK p55 subfamily member 2 | gi|156374008|ref|XP_001629601.1|predicted protein [Nematostella vectensis], 1e-289 |
| other nr_hits | gi|999986377|gb|KXJ25516.1|MAGUK p55 subfamily member 6 [Exaiptasia pallida] 2e-238; gi|1126188276|ref|XP_019626945.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X21 [Branchiostoma belcheri] 2e-176; gi|1126188278|ref|XP_019626946.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X22 [Branchiostoma belcheri] 2e-175; gi|1126188274|ref|XP_019626944.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X20 [Branchiostoma belcheri] 1e-172; gi|1126188272|ref|XP_019626943.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X19 [Branchiostoma belcheri] 2e-172; gi|1126188270|ref|XP_019626942.1|PREDICTED: MAGUK p55 subfamily member 6-like isoform X18 [Branchiostoma belcheri] 8e-170; gi|557016585|ref|XP_006008722.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X1 [Latimeria chalumnae] 1e-162; gi|557016588|ref|XP_006008723.1|PREDICTED: MAGUK p55 subfamily member 6 isoform X2 [Latimeria chalumnae] 1e-162; gi|530624900|ref|XP_005301999.1|PREDICTED: MAGUK p55 subfamily member 2 isoform X1 [Chrysemys picta bellii] 7e-161; | ||||||
| NvERTx.4.3597 | R-7 | 0.999973737681 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.106722 | R-7 | 0.999973737681 | E-5 | 1.0 | Q60809 | CCR4-NOT transcription complex subunit 7 | gi|156372834|ref|XP_001629240.1|predicted protein [Nematostella vectensis], 2e-151 |
| other nr_hits | gi|999983718|gb|KXJ22859.1|CCR4-NOT transcription complex subunit 7 [Exaiptasia pallida] 2e-138; gi|1005460530|ref|XP_015767349.1|PREDICTED: CCR4-NOT transcription complex subunit 7-like [Acropora digitifera] 4e-125; gi|919102308|ref|XP_013387545.1|PREDICTED: CCR4-NOT transcription complex subunit 7-like [Lingula anatina] 8e-112; gi|443688301|gb|ELT91034.1|hypothetical protein CAPTEDRAFT_96823 [Capitella teleta] 8e-112; gi|762116676|ref|XP_011441179.1|PREDICTED: CCR4-NOT transcription complex subunit 7 [Crassostrea gigas] 1e-110; gi|646712238|gb|KDR17086.1|CCR4-NOT transcription complex subunit 7 [Zootermopsis nevadensis] 3e-110; gi|665800502|ref|XP_008548219.1|PREDICTED: CCR4-NOT transcription complex subunit 7 isoform X1 [Microplitis demolitor] 4e-110; gi|665800506|ref|XP_008548221.1|PREDICTED: CCR4-NOT transcription complex subunit 7 isoform X3 [Microplitis demolitor] 4e-110; gi|665800504|ref|XP_008548220.1|PREDICTED: CCR4-NOT transcription complex subunit 7 isoform X2 [Microplitis demolitor] 4e-110; | ||||||
| NvERTx.4.114157 | R-7 | 0.999883671751 | E-5 | 1.0 | O89086 | RNA-binding protein 3 | gi|999971420|gb|KXJ10594.1|Transcription factor HES-1-A [Exaiptasia pallida], 2e-36 |
| other nr_hits | gi|156367055|ref|XP_001627235.1|predicted protein [Nematostella vectensis] 7e-34; gi|156336335|ref|XP_001619697.1|hypothetical protein NEMVEDRAFT_v1g248837 [Nematostella vectensis] 3e-31; gi|1005424498|ref|XP_015776410.1|PREDICTED: RNA-binding motif protein, X chromosome-like [Acropora digitifera] 2e-17; gi|837811121|ref|XP_012783216.1|PREDICTED: RNA-binding protein 3 [Ochotona princeps] 3e-11; gi|532107947|ref|XP_005338703.1|PREDICTED: RNA-binding protein 3 isoform X1 [Ictidomys tridecemlineatus] 3e-11; gi|532107949|ref|XP_005338704.1|PREDICTED: RNA-binding protein 3 isoform X2 [Ictidomys tridecemlineatus] 3e-11; gi|551604668|ref|XP_005783793.1|hypothetical protein EMIHUDRAFT_72362, partial [Emiliania huxleyi CCMP1516] 3e-11; gi|970736616|ref|XP_015102790.1|PREDICTED: RNA-binding protein 3 isoform X2 [Vicugna pacos] 5e-11; gi|744614479|ref|XP_010995176.1|PREDICTED: putative RNA-binding protein 3 isoform X2 [Camelus dromedarius] 5e-11; | ||||||
| NvERTx.4.114158 | R-7 | 0.999883671751 | E-5 | 1.0 | O89086 | RNA-binding protein 3 | gi|999971420|gb|KXJ10594.1|Transcription factor HES-1-A [Exaiptasia pallida], 2e-36 |
| other nr_hits | gi|156367055|ref|XP_001627235.1|predicted protein [Nematostella vectensis] 7e-34; gi|156336335|ref|XP_001619697.1|hypothetical protein NEMVEDRAFT_v1g248837 [Nematostella vectensis] 2e-31; gi|1005424498|ref|XP_015776410.1|PREDICTED: RNA-binding motif protein, X chromosome-like [Acropora digitifera] 2e-17; gi|837811121|ref|XP_012783216.1|PREDICTED: RNA-binding protein 3 [Ochotona princeps] 3e-11; gi|532107947|ref|XP_005338703.1|PREDICTED: RNA-binding protein 3 isoform X1 [Ictidomys tridecemlineatus] 3e-11; gi|532107949|ref|XP_005338704.1|PREDICTED: RNA-binding protein 3 isoform X2 [Ictidomys tridecemlineatus] 3e-11; gi|551604668|ref|XP_005783793.1|hypothetical protein EMIHUDRAFT_72362, partial [Emiliania huxleyi CCMP1516] 3e-11; gi|970736616|ref|XP_015102790.1|PREDICTED: RNA-binding protein 3 isoform X2 [Vicugna pacos] 5e-11; gi|744614479|ref|XP_010995176.1|PREDICTED: putative RNA-binding protein 3 isoform X2 [Camelus dromedarius] 5e-11; | ||||||
| NvERTx.4.114161 | R-7 | 0.999883671751 | E-5 | 1.0 | O89086 | RNA-binding protein 3 | gi|999971420|gb|KXJ10594.1|Transcription factor HES-1-A [Exaiptasia pallida], 1e-36 |
| other nr_hits | gi|156367055|ref|XP_001627235.1|predicted protein [Nematostella vectensis] 7e-34; gi|156336335|ref|XP_001619697.1|hypothetical protein NEMVEDRAFT_v1g248837 [Nematostella vectensis] 2e-31; gi|1005424498|ref|XP_015776410.1|PREDICTED: RNA-binding motif protein, X chromosome-like [Acropora digitifera] 2e-17; gi|837811121|ref|XP_012783216.1|PREDICTED: RNA-binding protein 3 [Ochotona princeps] 3e-11; gi|532107947|ref|XP_005338703.1|PREDICTED: RNA-binding protein 3 isoform X1 [Ictidomys tridecemlineatus] 3e-11; gi|532107949|ref|XP_005338704.1|PREDICTED: RNA-binding protein 3 isoform X2 [Ictidomys tridecemlineatus] 3e-11; gi|551604668|ref|XP_005783793.1|hypothetical protein EMIHUDRAFT_72362, partial [Emiliania huxleyi CCMP1516] 3e-11; gi|970736616|ref|XP_015102790.1|PREDICTED: RNA-binding protein 3 isoform X2 [Vicugna pacos] 5e-11; gi|744614479|ref|XP_010995176.1|PREDICTED: putative RNA-binding protein 3 isoform X2 [Camelus dromedarius] 5e-11; | ||||||
| NvERTx.4.131179 | R-6 | 0.998238799649 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.131180 | R-6 | 0.998238799649 | E-5 | 1.0 | Q9WTP0 | Band 4.1-like protein 1 | gi|156351016|ref|XP_001622324.1|hypothetical protein NEMVEDRAFT_v1g176215 [Nematostella vectensis], 1e-227 |
| other nr_hits | gi|632939744|ref|XP_007883002.1|PREDICTED: band 4.1-like protein 1 [Callorhinchus milii] 2e-85; gi|1126210382|ref|XP_019638945.1|PREDICTED: protein 4.1-like isoform X6 [Branchiostoma belcheri] 2e-82; gi|1126210420|ref|XP_019638964.1|PREDICTED: protein 4.1-like isoform X25 [Branchiostoma belcheri] 2e-82; gi|1126210416|ref|XP_019638962.1|PREDICTED: protein 4.1-like isoform X23 [Branchiostoma belcheri] 2e-82; gi|1158851122|ref|XP_020325334.1|band 4.1-like protein 1 [Oncorhynchus kisutch] 5e-82; gi|1158869746|ref|XP_020338658.1|band 4.1-like protein 1 isoform X3 [Oncorhynchus kisutch] 3e-81; gi|1158869744|ref|XP_020338657.1|band 4.1-like protein 1 isoform X2 [Oncorhynchus kisutch] 3e-81; gi|1158869740|ref|XP_020338655.1|band 4.1-like protein 1 isoform X1 [Oncorhynchus kisutch] 3e-81; gi|1158869750|ref|XP_020338660.1|band 4.1-like protein 1 isoform X5 [Oncorhynchus kisutch] 3e-81; | ||||||
| NvERTx.4.131181 | R-6 | 0.998238799649 | E-5 | 1.0 | Q9WTP0 | Band 4.1-like protein 1 | gi|156351016|ref|XP_001622324.1|hypothetical protein NEMVEDRAFT_v1g176215 [Nematostella vectensis], 1e-227 |
| other nr_hits | gi|632939744|ref|XP_007883002.1|PREDICTED: band 4.1-like protein 1 [Callorhinchus milii] 2e-85; gi|1126210382|ref|XP_019638945.1|PREDICTED: protein 4.1-like isoform X6 [Branchiostoma belcheri] 2e-82; gi|1126210420|ref|XP_019638964.1|PREDICTED: protein 4.1-like isoform X25 [Branchiostoma belcheri] 2e-82; gi|1126210416|ref|XP_019638962.1|PREDICTED: protein 4.1-like isoform X23 [Branchiostoma belcheri] 2e-82; gi|1158851122|ref|XP_020325334.1|band 4.1-like protein 1 [Oncorhynchus kisutch] 5e-82; gi|1158869746|ref|XP_020338658.1|band 4.1-like protein 1 isoform X3 [Oncorhynchus kisutch] 3e-81; gi|1158869744|ref|XP_020338657.1|band 4.1-like protein 1 isoform X2 [Oncorhynchus kisutch] 3e-81; gi|1158869740|ref|XP_020338655.1|band 4.1-like protein 1 isoform X1 [Oncorhynchus kisutch] 3e-81; gi|1158869750|ref|XP_020338660.1|band 4.1-like protein 1 isoform X5 [Oncorhynchus kisutch] 3e-81; | ||||||
| NvERTx.4.131182 | R-6 | 0.998238799649 | E-5 | 1.0 | Q9WTP0 | Band 4.1-like protein 1 | gi|156351016|ref|XP_001622324.1|hypothetical protein NEMVEDRAFT_v1g176215 [Nematostella vectensis], 2e-227 |
| other nr_hits | gi|632939744|ref|XP_007883002.1|PREDICTED: band 4.1-like protein 1 [Callorhinchus milii] 2e-85; gi|1126210382|ref|XP_019638945.1|PREDICTED: protein 4.1-like isoform X6 [Branchiostoma belcheri] 2e-82; gi|1126210420|ref|XP_019638964.1|PREDICTED: protein 4.1-like isoform X25 [Branchiostoma belcheri] 2e-82; gi|1126210416|ref|XP_019638962.1|PREDICTED: protein 4.1-like isoform X23 [Branchiostoma belcheri] 2e-82; gi|1158851122|ref|XP_020325334.1|band 4.1-like protein 1 [Oncorhynchus kisutch] 4e-82; gi|1083282320|ref|XP_018589624.1|PREDICTED: band 4.1-like protein 1 isoform X1 [Scleropages formosus] 1e-81; gi|1083282324|ref|XP_018589626.1|PREDICTED: band 4.1-like protein 1 isoform X3 [Scleropages formosus] 2e-81; gi|1158869746|ref|XP_020338658.1|band 4.1-like protein 1 isoform X3 [Oncorhynchus kisutch] 2e-81; gi|1158869744|ref|XP_020338657.1|band 4.1-like protein 1 isoform X2 [Oncorhynchus kisutch] 2e-81; | ||||||
| NvERTx.4.131183 | R-6 | 0.998238799649 | E-5 | 1.0 | Q9WTP0 | Band 4.1-like protein 1 | gi|156351016|ref|XP_001622324.1|hypothetical protein NEMVEDRAFT_v1g176215 [Nematostella vectensis], 1e-227 |
| other nr_hits | gi|632939744|ref|XP_007883002.1|PREDICTED: band 4.1-like protein 1 [Callorhinchus milii] 2e-85; gi|1126210382|ref|XP_019638945.1|PREDICTED: protein 4.1-like isoform X6 [Branchiostoma belcheri] 2e-82; gi|1126210420|ref|XP_019638964.1|PREDICTED: protein 4.1-like isoform X25 [Branchiostoma belcheri] 2e-82; gi|1126210416|ref|XP_019638962.1|PREDICTED: protein 4.1-like isoform X23 [Branchiostoma belcheri] 2e-82; gi|1158851122|ref|XP_020325334.1|band 4.1-like protein 1 [Oncorhynchus kisutch] 4e-82; gi|1158869746|ref|XP_020338658.1|band 4.1-like protein 1 isoform X3 [Oncorhynchus kisutch] 3e-81; gi|1158869744|ref|XP_020338657.1|band 4.1-like protein 1 isoform X2 [Oncorhynchus kisutch] 3e-81; gi|1158869740|ref|XP_020338655.1|band 4.1-like protein 1 isoform X1 [Oncorhynchus kisutch] 3e-81; gi|1158869750|ref|XP_020338660.1|band 4.1-like protein 1 isoform X5 [Oncorhynchus kisutch] 3e-81; | ||||||
| NvERTx.4.162334 | R-7 | 0.999962089371 | E-5 | 1.0 | Q6NYV9 | RNA polymerase-associated protein LEO1 | gi|156394980|ref|XP_001636890.1|predicted protein [Nematostella vectensis], 4e-146 |
| other nr_hits | gi|999984132|gb|KXJ23273.1|RNA polymerase-associated protein LEO1 [Exaiptasia pallida] 7e-89; gi|1005470725|ref|XP_015772244.1|PREDICTED: RNA polymerase-associated protein LEO1-like [Acropora digitifera] 4e-85; gi|1069338617|ref|XP_018108564.1|PREDICTED: RNA polymerase-associated protein LEO1-like [Xenopus laevis] 9e-65; gi|1101630796|ref|XP_018978896.1|PREDICTED: RNA polymerase-associated protein LEO1 [Cyprinus carpio] 1e-64; gi|1050384749|gb|OCT86943.1|hypothetical protein XELAEV_18020633mg [Xenopus laevis] 1e-64; gi|147900849|ref|NP_001085280.1|RNA polymerase-associated protein LEO1 [Xenopus laevis] 1e-64; gi|221119154|ref|XP_002158694.1|PREDICTED: RNA polymerase-associated protein LEO1-like [Hydra vulgaris] 2e-64; gi|602630651|ref|XP_007422239.1|PREDICTED: RNA polymerase-associated protein LEO1 [Python bivittatus] 2e-64; gi|512828164|ref|XP_002936391.2|PREDICTED: RNA polymerase-associated protein LEO1 [Xenopus tropicalis] 2e-64; | ||||||
| NvERTx.4.162335 | R-7 | 0.999962089371 | E-5 | 1.0 | Q6NYV9 | RNA polymerase-associated protein LEO1 | gi|156394980|ref|XP_001636890.1|predicted protein [Nematostella vectensis], 7e-151 |
| other nr_hits | gi|999984132|gb|KXJ23273.1|RNA polymerase-associated protein LEO1 [Exaiptasia pallida] 8e-89; gi|1005470725|ref|XP_015772244.1|PREDICTED: RNA polymerase-associated protein LEO1-like [Acropora digitifera] 4e-85; gi|1069338617|ref|XP_018108564.1|PREDICTED: RNA polymerase-associated protein LEO1-like [Xenopus laevis] 1e-64; gi|1101630796|ref|XP_018978896.1|PREDICTED: RNA polymerase-associated protein LEO1 [Cyprinus carpio] 1e-64; gi|1050384749|gb|OCT86943.1|hypothetical protein XELAEV_18020633mg [Xenopus laevis] 1e-64; gi|147900849|ref|NP_001085280.1|RNA polymerase-associated protein LEO1 [Xenopus laevis] 1e-64; gi|221119154|ref|XP_002158694.1|PREDICTED: RNA polymerase-associated protein LEO1-like [Hydra vulgaris] 2e-64; gi|602630651|ref|XP_007422239.1|PREDICTED: RNA polymerase-associated protein LEO1 [Python bivittatus] 2e-64; gi|512828164|ref|XP_002936391.2|PREDICTED: RNA polymerase-associated protein LEO1 [Xenopus tropicalis] 2e-64; | ||||||
| NvERTx.4.162336 | R-7 | 0.999962089371 | E-5 | 1.0 | Q6NYV9 | RNA polymerase-associated protein LEO1 | gi|156394980|ref|XP_001636890.1|predicted protein [Nematostella vectensis], 3e-20 |
| other nr_hits | gi|1005470725|ref|XP_015772244.1|PREDICTED: RNA polymerase-associated protein LEO1-like [Acropora digitifera] 5e-11; gi|632938249|ref|XP_007904284.1|PREDICTED: RNA polymerase-associated protein LEO1 [Callorhinchus milii] 8e-09; gi|1073720056|gb|JAU02956.1|putative rna polymerase-associated protein leo1, partial [Amblyomma sculptum] 1e-08; gi|762159179|ref|XP_011417789.1|PREDICTED: RNA polymerase-associated protein LEO1 [Crassostrea gigas] 1e-08; gi|405960501|gb|EKC26422.1|RNA polymerase-associated protein LEO1 [Crassostrea gigas] 1e-08; gi|1007737914|ref|XP_015809398.1|PREDICTED: RNA polymerase-associated protein LEO1 [Nothobranchius furzeri] 2e-08; gi|974067209|ref|XP_015231078.1|PREDICTED: RNA polymerase-associated protein LEO1 [Cyprinodon variegatus] 2e-08; gi|961861602|ref|XP_014845730.1|PREDICTED: RNA polymerase-associated protein LEO1 isoform X1 [Poecilia mexicana] 2e-08; gi|941780480|ref|XP_014324491.1|PREDICTED: RNA polymerase-associated protein LEO1, partial [Xiphophorus maculatus] 2e-08; | ||||||
| NvERTx.4.166758 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|811389931|gb|AKE25951.1|sarco/endoplasmic reticulum calcium ATPase [Hyriopsis cumingii] 0e+00; gi|780010615|ref|XP_011663710.1|PREDICTED: sarco/endoplasmic reticulum calcium transporting ATPase isoform X1 [Strongylocentrotus purpuratus] 0e+00; gi|919080717|ref|XP_013421498.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X1 [Lingula anatina] 0e+00; gi|919080727|ref|XP_013421503.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X4 [Lingula anatina] 0e+00; gi|919080732|ref|XP_013421505.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X5 [Lingula anatina] 0e+00; gi|919080735|ref|XP_013421506.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X6 [Lingula anatina] 0e+00; gi|152003983|gb|ABS19815.1|sarco/endoplasmic reticulum calcium ATPase isoform A [Pinctada fucata] 0e+00; gi|152003985|gb|ABS19816.1|sarco/endoplasmic reticulum calcium ATPase isoform B [Pinctada fucata] 0e+00; gi|152003987|gb|ABS19817.1|sarco/endoplasmic reticulum calcium ATPase isoform C [Pinctada fucata] 0e+00; | ||||||
| NvERTx.4.166767 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 0e+00; gi|919080717|ref|XP_013421498.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X1 [Lingula anatina] 0e+00; gi|919080727|ref|XP_013421503.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X4 [Lingula anatina] 0e+00; gi|919080732|ref|XP_013421505.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X5 [Lingula anatina] 0e+00; gi|919080735|ref|XP_013421506.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X6 [Lingula anatina] 0e+00; gi|811389931|gb|AKE25951.1|sarco/endoplasmic reticulum calcium ATPase [Hyriopsis cumingii] 0e+00; gi|919080722|ref|XP_013421500.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X3 [Lingula anatina] 0e+00; gi|780010615|ref|XP_011663710.1|PREDICTED: sarco/endoplasmic reticulum calcium transporting ATPase isoform X1 [Strongylocentrotus purpuratus] 0e+00; gi|919080720|ref|XP_013421499.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X2 [Lingula anatina] 0e+00; | ||||||
| NvERTx.4.166768 | R-7 | 0.959678339349 | E-5 | 1.0 | Q9YGL9 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 5e-226 |
| other nr_hits | gi|1005434781|ref|XP_015754893.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Acropora digitifera] 3e-189; gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 2e-181; gi|1087803684|ref|XP_009861043.2|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2-like isoform X2 [Ciona intestinalis] 2e-172; gi|1087803682|ref|XP_009861042.2|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Ciona intestinalis] 2e-172; gi|585690683|ref|XP_006821107.1|PREDICTED: LOW QUALITY PROTEIN: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Saccoglossus kowalevskii] 8e-172; gi|1126158091|ref|XP_019634652.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X2 [Branchiostoma belcheri] 4e-171; gi|1073703943|gb|JAT95028.1|putative calcium-transporting atpase sarcoplasmic/endoplasmic reticulum type calcium pump isoform 1 [Amblyomma aureolatum] 2e-170; gi|1126158089|ref|XP_019634586.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Branchiostoma belcheri] 8e-170; gi|1126158093|ref|XP_019634739.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X3 [Branchiostoma belcheri] 8e-170; | ||||||
| NvERTx.4.166769 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 9e-27 |
| other nr_hits | gi|478261155|gb|ENN80688.1|hypothetical protein YQE_02890, partial [Dendroctonus ponderosae] 3e-16; gi|546674790|gb|ERL86086.1|hypothetical protein D910_03500 [Dendroctonus ponderosae] 9e-16; gi|811389931|gb|AKE25951.1|sarco/endoplasmic reticulum calcium ATPase [Hyriopsis cumingii] 2e-15; gi|1130209790|ref|XP_019754226.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type [Dendroctonus ponderosae] 3e-15; gi|1073703943|gb|JAT95028.1|putative calcium-transporting atpase sarcoplasmic/endoplasmic reticulum type calcium pump isoform 1 [Amblyomma aureolatum] 3e-15; gi|260181324|gb|ACX35338.1|calcium-transporting ATPase variant 1 [Lumbricus rubellus] 4e-15; gi|260181326|gb|ACX35339.1|calcium-transporting ATPase variant 2 [Lumbricus rubellus] 4e-15; gi|1101366064|ref|XP_018914247.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type isoform X1 [Bemisia tabaci] 6e-15; gi|1101366070|ref|XP_018914251.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type isoform X2 [Bemisia tabaci] 6e-15; | ||||||
| NvERTx.4.166770 | R-7 | 0.959678339349 | E-5 | 1.0 | Q64578 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 4e-108 |
| other nr_hits | gi|1005434781|ref|XP_015754893.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Acropora digitifera] 1e-88; gi|1126158089|ref|XP_019634586.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Branchiostoma belcheri] 6e-82; gi|1126158093|ref|XP_019634739.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X3 [Branchiostoma belcheri] 6e-82; gi|1126158091|ref|XP_019634652.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X2 [Branchiostoma belcheri] 6e-82; gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 1e-81; gi|811389931|gb|AKE25951.1|sarco/endoplasmic reticulum calcium ATPase [Hyriopsis cumingii] 1e-81; gi|1083296699|ref|XP_018597395.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 3, partial [Scleropages formosus] 3e-81; gi|260181324|gb|ACX35338.1|calcium-transporting ATPase variant 1 [Lumbricus rubellus] 3e-81; gi|260181326|gb|ACX35339.1|calcium-transporting ATPase variant 2 [Lumbricus rubellus] 3e-81; | ||||||
| NvERTx.4.166772 | R-7 | 0.959678339349 | E-5 | 1.0 | Q03669 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 1e-38 |
| other nr_hits | gi|1005434781|ref|XP_015754893.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Acropora digitifera] 2e-34; gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 1e-33; gi|326519737|dbj|BAK00241.1|predicted protein [Hordeum vulgare subsp. vulgare] 3e-33; gi|1121915801|ref|XP_019476563.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X1 [Meleagris gallopavo] 5e-33; gi|1121850637|ref|XP_019394241.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X1 [Crocodylus porosus] 5e-33; gi|1121850642|ref|XP_019394242.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X2 [Crocodylus porosus] 5e-33; gi|1117279653|ref|XP_019362862.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X1 [Gavialis gangeticus] 5e-33; gi|1117279656|ref|XP_019362863.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X2 [Gavialis gangeticus] 5e-33; gi|1083296699|ref|XP_018597395.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 3, partial [Scleropages formosus] 5e-33; | ||||||
| NvERTx.4.166759 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|780010615|ref|XP_011663710.1|PREDICTED: sarco/endoplasmic reticulum calcium transporting ATPase isoform X1 [Strongylocentrotus purpuratus] 0e+00; gi|83165280|ref|NP_001032719.1|sarco/endoplasmic reticulum calcium transporting ATPase [Strongylocentrotus purpuratus] 0e+00; gi|919080717|ref|XP_013421498.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X1 [Lingula anatina] 0e+00; gi|152003983|gb|ABS19815.1|sarco/endoplasmic reticulum calcium ATPase isoform A [Pinctada fucata] 0e+00; gi|152003985|gb|ABS19816.1|sarco/endoplasmic reticulum calcium ATPase isoform B [Pinctada fucata] 0e+00; gi|152003987|gb|ABS19817.1|sarco/endoplasmic reticulum calcium ATPase isoform C [Pinctada fucata] 0e+00; gi|919080727|ref|XP_013421503.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X4 [Lingula anatina] 0e+00; gi|919080732|ref|XP_013421505.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X5 [Lingula anatina] 0e+00; gi|919080735|ref|XP_013421506.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X6 [Lingula anatina] 0e+00; | ||||||
| NvERTx.4.166760 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 9e-27 |
| other nr_hits | gi|478261155|gb|ENN80688.1|hypothetical protein YQE_02890, partial [Dendroctonus ponderosae] 3e-16; gi|546674790|gb|ERL86086.1|hypothetical protein D910_03500 [Dendroctonus ponderosae] 9e-16; gi|811389931|gb|AKE25951.1|sarco/endoplasmic reticulum calcium ATPase [Hyriopsis cumingii] 2e-15; gi|1130209790|ref|XP_019754226.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type [Dendroctonus ponderosae] 3e-15; gi|1073703943|gb|JAT95028.1|putative calcium-transporting atpase sarcoplasmic/endoplasmic reticulum type calcium pump isoform 1 [Amblyomma aureolatum] 3e-15; gi|260181324|gb|ACX35338.1|calcium-transporting ATPase variant 1 [Lumbricus rubellus] 4e-15; gi|260181326|gb|ACX35339.1|calcium-transporting ATPase variant 2 [Lumbricus rubellus] 4e-15; gi|1101366064|ref|XP_018914247.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type isoform X1 [Bemisia tabaci] 6e-15; gi|1101366070|ref|XP_018914251.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type isoform X2 [Bemisia tabaci] 6e-15; | ||||||
| NvERTx.4.166761 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 4e-297; gi|919080717|ref|XP_013421498.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X1 [Lingula anatina] 3e-294; gi|919080720|ref|XP_013421499.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X2 [Lingula anatina] 3e-294; gi|919080722|ref|XP_013421500.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X3 [Lingula anatina] 3e-294; gi|919080727|ref|XP_013421503.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X4 [Lingula anatina] 3e-294; gi|919080732|ref|XP_013421505.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X5 [Lingula anatina] 3e-294; gi|919080735|ref|XP_013421506.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X6 [Lingula anatina] 3e-294; gi|1087803684|ref|XP_009861043.2|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2-like isoform X2 [Ciona intestinalis] 3e-293; gi|1087803682|ref|XP_009861042.2|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Ciona intestinalis] 3e-293; | ||||||
| NvERTx.4.166762 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 4e-297; gi|919080717|ref|XP_013421498.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X1 [Lingula anatina] 3e-294; gi|919080720|ref|XP_013421499.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X2 [Lingula anatina] 3e-294; gi|919080722|ref|XP_013421500.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X3 [Lingula anatina] 3e-294; gi|919080727|ref|XP_013421503.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X4 [Lingula anatina] 3e-294; gi|919080732|ref|XP_013421505.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X5 [Lingula anatina] 3e-294; gi|919080735|ref|XP_013421506.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like isoform X6 [Lingula anatina] 3e-294; gi|1087803684|ref|XP_009861043.2|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2-like isoform X2 [Ciona intestinalis] 3e-293; gi|1087803682|ref|XP_009861042.2|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Ciona intestinalis] 3e-293; | ||||||
| NvERTx.4.166763 | R-7 | 0.959678339349 | E-5 | 1.0 | Q64578 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 7e-143 |
| other nr_hits | gi|1005434781|ref|XP_015754893.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Acropora digitifera] 7e-119; gi|1126158091|ref|XP_019634652.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X2 [Branchiostoma belcheri] 3e-112; gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 5e-112; gi|1126158089|ref|XP_019634586.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Branchiostoma belcheri] 6e-111; gi|1126158093|ref|XP_019634739.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X3 [Branchiostoma belcheri] 6e-111; gi|1072275947|ref|XP_018423924.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X1 [Nanorana parkeri] 2e-110; gi|1072275951|ref|XP_018423926.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X3 [Nanorana parkeri] 2e-110; gi|811389931|gb|AKE25951.1|sarco/endoplasmic reticulum calcium ATPase [Hyriopsis cumingii] 2e-110; gi|831546332|ref|XP_012726552.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X1 [Fundulus heteroclitus] 2e-110; | ||||||
| NvERTx.4.166764 | R-7 | 0.959678339349 | E-5 | 1.0 | Q03669 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 1e-14 |
| other nr_hits | gi|1083296699|ref|XP_018597395.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 3, partial [Scleropages formosus] 8e-14; gi|1025165035|ref|XP_016418779.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Sinocyclocheilus rhinocerous] 1e-13; gi|1025165040|ref|XP_016418780.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X2 [Sinocyclocheilus rhinocerous] 1e-13; gi|1025165052|ref|XP_016418782.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2-like isoform X4 [Sinocyclocheilus rhinocerous] 1e-13; gi|1024977815|ref|XP_016310453.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Sinocyclocheilus anshuiensis] 1e-13; gi|1024977819|ref|XP_016310455.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X3 [Sinocyclocheilus anshuiensis] 1e-13; gi|1020422996|ref|XP_016130605.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Sinocyclocheilus grahami] 1e-13; gi|1020423000|ref|XP_016130607.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X3 [Sinocyclocheilus grahami] 1e-13; gi|1020423002|ref|XP_016130609.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 3-like isoform X4 [Sinocyclocheilus grahami] 1e-13; | ||||||
| NvERTx.4.166765 | R-7 | 0.959678339349 | E-5 | 1.0 | Q8R429 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 4e-78 |
| other nr_hits | gi|1005434781|ref|XP_015754893.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Acropora digitifera] 3e-62; gi|1083296699|ref|XP_018597395.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 3, partial [Scleropages formosus] 2e-57; gi|974115841|ref|XP_015257355.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2-like [Cyprinodon variegatus] 2e-57; gi|1143368368|ref|XP_019937612.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 [Paralichthys olivaceus] 4e-57; gi|961999596|ref|XP_014828327.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X1 [Poecilia mexicana] 4e-57; gi|961999600|ref|XP_014828328.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X2 [Poecilia mexicana] 4e-57; gi|962010057|ref|XP_014878645.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X2 [Poecilia latipinna] 4e-57; gi|831546332|ref|XP_012726552.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X1 [Fundulus heteroclitus] 4e-57; gi|831546335|ref|XP_012726553.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 2 isoform X2 [Fundulus heteroclitus] 4e-57; | ||||||
| NvERTx.4.166766 | R-7 | 0.959678339349 | E-5 | 1.0 | Q9YGL9 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 4e-204 |
| other nr_hits | gi|1005434781|ref|XP_015754893.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Acropora digitifera] 4e-174; gi|999972104|gb|KXJ11278.1|Sarcoplasmic/endoplasmic reticulum calcium ATPase 3 [Exaiptasia pallida] 5e-167; gi|1126158091|ref|XP_019634652.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X2 [Branchiostoma belcheri] 2e-161; gi|1126158089|ref|XP_019634586.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X1 [Branchiostoma belcheri] 4e-160; gi|1126158093|ref|XP_019634739.1|PREDICTED: sarcoplasmic/endoplasmic reticulum calcium ATPase 1-like isoform X3 [Branchiostoma belcheri] 4e-160; gi|1073703943|gb|JAT95028.1|putative calcium-transporting atpase sarcoplasmic/endoplasmic reticulum type calcium pump isoform 1 [Amblyomma aureolatum] 6e-160; gi|585690683|ref|XP_006821107.1|PREDICTED: LOW QUALITY PROTEIN: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Saccoglossus kowalevskii] 7e-159; gi|1069817852|ref|XP_018332133.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type isoform X1 [Agrilus planipennis] 2e-158; gi|1069817859|ref|XP_018332136.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type isoform X2 [Agrilus planipennis] 2e-158; | ||||||
| NvERTx.4.166773 | R-7 | 0.959678339349 | E-5 | 1.0 | Q7PPA5 | Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type | gi|156402299|ref|XP_001639528.1|predicted protein [Nematostella vectensis], 9e-37 |
| other nr_hits | gi|152003983|gb|ABS19815.1|sarco/endoplasmic reticulum calcium ATPase isoform A [Pinctada fucata] 2e-25; gi|152003985|gb|ABS19816.1|sarco/endoplasmic reticulum calcium ATPase isoform B [Pinctada fucata] 2e-25; gi|152003987|gb|ABS19817.1|sarco/endoplasmic reticulum calcium ATPase isoform C [Pinctada fucata] 2e-25; gi|811389931|gb|AKE25951.1|sarco/endoplasmic reticulum calcium ATPase [Hyriopsis cumingii] 3e-25; gi|443717277|gb|ELU08428.1|hypothetical protein CAPTEDRAFT_165811 [Capitella teleta] 5e-25; gi|56757035|gb|AAW26689.1|SJCHGC05468 protein [Schistosoma japonicum] 5e-25; gi|918290408|gb|KOF68930.1|hypothetical protein OCBIM_22006279mg [Octopus bimaculoides] 1e-24; gi|844872365|ref|XP_012800781.1|Calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type [Schistosoma haematobium] 1e-24; gi|957821886|ref|XP_014681278.1|PREDICTED: calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type-like [Priapulus caudatus] 1e-24; | ||||||
| NvERTx.4.183149 | R-6 | 0.993474632239 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.183150 | R-6 | 0.993474632239 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.183151 | R-6 | 0.993474632239 | E-5 | 1.0 | Q9CQC9 | GTP-binding protein SAR1b | gi|156397201|ref|XP_001637780.1|predicted protein [Nematostella vectensis], 7e-108 |
| other nr_hits | gi|999982088|gb|KXJ21240.1|GTP-binding protein SAR1b [Exaiptasia pallida] 4e-97; gi|1069792540|ref|XP_018321337.1|PREDICTED: GTP-binding protein SAR1b [Agrilus planipennis] 7e-86; gi|936695465|ref|XP_014225714.1|PREDICTED: GTP-binding protein SAR1b [Trichogramma pretiosum] 7e-86; gi|1133421312|ref|XP_019866277.1|PREDICTED: GTP-binding protein SAR1b [Aethina tumida] 2e-85; gi|443721469|gb|ELU10760.1|hypothetical protein CAPTEDRAFT_156190 [Capitella teleta] 2e-85; gi|676480292|ref|XP_009062014.1|hypothetical protein LOTGIDRAFT_210455 [Lottia gigantea] 3e-85; gi|1080063541|ref|XP_018573862.1|PREDICTED: GTP-binding protein SAR1b isoform X1 [Anoplophora glabripennis] 4e-85; gi|1000716784|ref|XP_015590503.1|PREDICTED: GTP-binding protein SAR1 isoform X2 [Cephus cinctus] 5e-85; gi|156547852|ref|XP_001605005.1|PREDICTED: GTP-binding protein SAR1 [Nasonia vitripennis] 5e-85; | ||||||
| NvERTx.4.198881 | R-4 | 0.831315721485 | E-5 | 1.0 | Q9H9Y6 | DNA-directed RNA polymerase I subunit RPA2 | gi|1005469033|ref|XP_015771434.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2-like [Acropora digitifera], 1e-39 |
| other nr_hits | gi|999980411|gb|KXJ19585.1|DNA-directed RNA polymerase I subunit RPA2 [Exaiptasia pallida] 1e-38; gi|1126204888|ref|XP_019635951.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2-like [Branchiostoma belcheri] 2e-34; gi|926625781|ref|XP_013778405.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2-like, partial [Limulus polyphemus] 1e-29; gi|591368307|ref|XP_007058998.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 [Chelonia mydas] 4e-28; gi|465984460|gb|EMP36557.1|DNA-directed RNA polymerase I subunit RPA2 [Chelonia mydas] 4e-28; gi|558221462|ref|XP_006136012.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 [Pelodiscus sinensis] 5e-28; gi|632952911|ref|XP_007892112.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 isoform X1 [Callorhinchus milii] 7e-28; gi|632952913|ref|XP_007892113.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 isoform X2 [Callorhinchus milii] 7e-28; gi|1121897029|ref|XP_019404522.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 [Crocodylus porosus] 9e-28; | ||||||
| NvERTx.4.198883 | R-4 | 0.831315721485 | E-5 | 1.0 | Q9H9Y6 | DNA-directed RNA polymerase I subunit RPA2 | gi|1005469033|ref|XP_015771434.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2-like [Acropora digitifera], 1e-39 |
| other nr_hits | gi|999980411|gb|KXJ19585.1|DNA-directed RNA polymerase I subunit RPA2 [Exaiptasia pallida] 1e-38; gi|1126204888|ref|XP_019635951.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2-like [Branchiostoma belcheri] 2e-34; gi|926625781|ref|XP_013778405.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2-like, partial [Limulus polyphemus] 1e-29; gi|591368307|ref|XP_007058998.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 [Chelonia mydas] 4e-28; gi|465984460|gb|EMP36557.1|DNA-directed RNA polymerase I subunit RPA2 [Chelonia mydas] 4e-28; gi|558221462|ref|XP_006136012.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 [Pelodiscus sinensis] 5e-28; gi|632952911|ref|XP_007892112.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 isoform X1 [Callorhinchus milii] 7e-28; gi|632952913|ref|XP_007892113.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 isoform X2 [Callorhinchus milii] 7e-28; gi|1121897029|ref|XP_019404522.1|PREDICTED: DNA-directed RNA polymerase I subunit RPA2 [Crocodylus porosus] 9e-28; | ||||||
| NvERTx.4.4052 | R-6 | 0.885434800107 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
