Explore Mfuzz Clusters
Mfuzz cluster
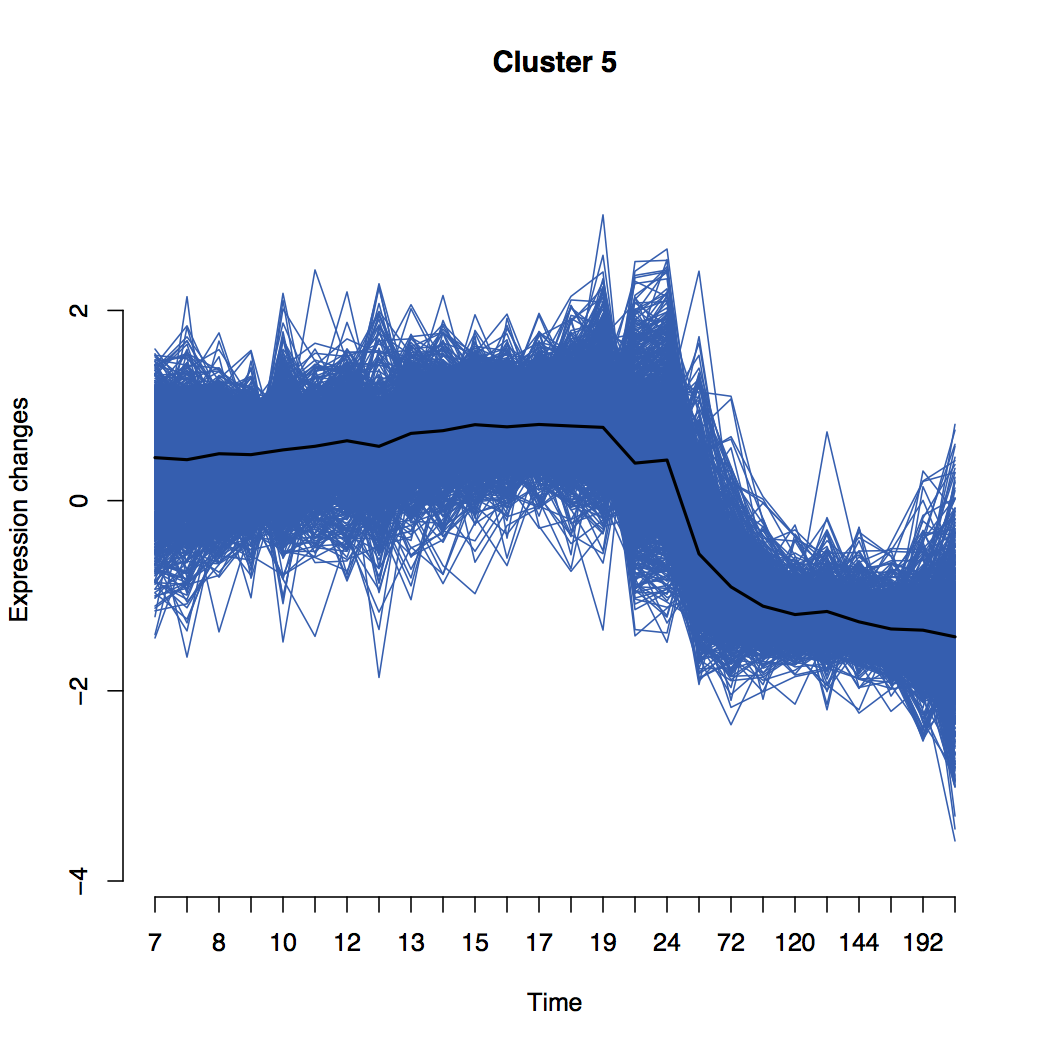
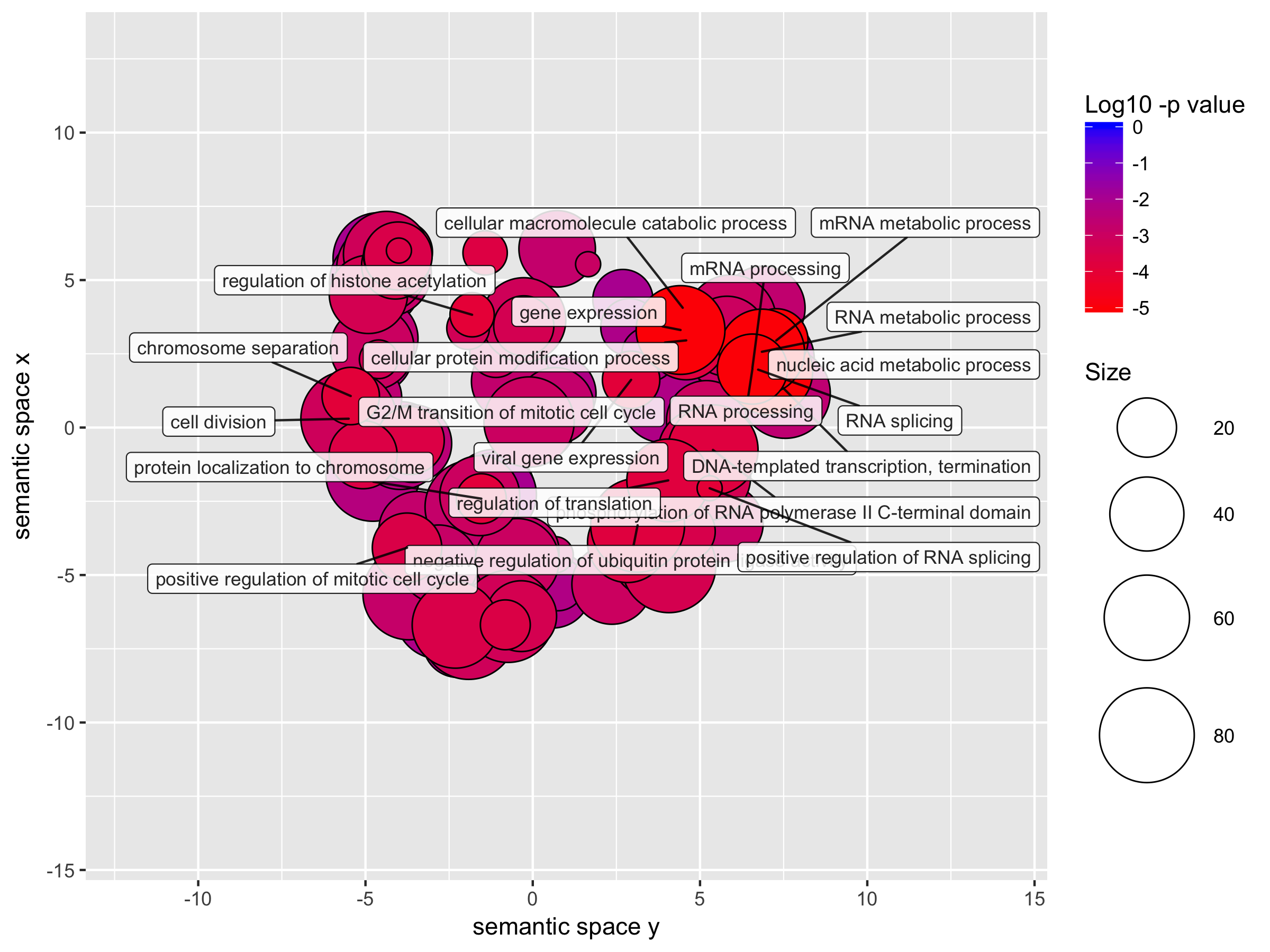
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.61659 | R-5 | 0.998050247064 | E-5 | 0.99999989788 | Q2TA25 | Serine/threonine-protein kinase PLK1 | gi|156375039|ref|XP_001629890.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999974374|gb|KXJ13548.1|Serine/threonine-protein kinase PLK1 [Exaiptasia pallida] 6e-226; gi|919040870|ref|XP_013403756.1|PREDICTED: serine/threonine-protein kinase PLK1-like [Lingula anatina] 4e-203; gi|1126166297|ref|XP_019614966.1|PREDICTED: serine/threonine-protein kinase PLK1-like [Branchiostoma belcheri] 5e-201; gi|762126579|ref|XP_011446350.1|PREDICTED: serine/threonine-protein kinase PLK1 [Crassostrea gigas] 2e-199; gi|334333140|ref|XP_001377703.2|PREDICTED: serine/threonine-protein kinase PLK1 [Monodelphis domestica] 3e-199; gi|443697887|gb|ELT98162.1|hypothetical protein CAPTEDRAFT_160517 [Capitella teleta] 4e-199; gi|957824411|ref|XP_014661948.1|PREDICTED: serine/threonine-protein kinase PLK1-like isoform X1 [Priapulus caudatus] 4e-199; gi|260792416|ref|XP_002591211.1|hypothetical protein BRAFLDRAFT_247541 [Branchiostoma floridae] 5e-199; gi|1147381311|ref|XP_020040036.1|serine/threonine-protein kinase PLK1 [Castor canadensis] 3e-198; | ||||||
| NvERTx.4.180031 | R-7 | 0.843924570274 | E-5 | 0.999999897296 | Q9WVT6 | Carbonic anhydrase 14 | gi|150456390|tpg|DAA06053.1|TPA_inf: carbonic anhydrase 1 [Nematostella vectensis], 8e-22 |
| other nr_hits | gi|156398335|ref|XP_001638144.1|predicted protein [Nematostella vectensis] 2e-15; | ||||||
| NvERTx.4.180032 | R-7 | 0.843924570274 | E-5 | 0.999999897296 | Q9WVT6 | Carbonic anhydrase 14 | gi|150456390|tpg|DAA06053.1|TPA_inf: carbonic anhydrase 1 [Nematostella vectensis], 8e-22 |
| other nr_hits | gi|156398335|ref|XP_001638144.1|predicted protein [Nematostella vectensis] 2e-15; | ||||||
| NvERTx.4.180036 | R-7 | 0.843924570274 | E-5 | 0.999999897296 | Q92051 | Carbonic anhydrase | gi|150456390|tpg|DAA06053.1|TPA_inf: carbonic anhydrase 1 [Nematostella vectensis], 1e-155 |
| other nr_hits | gi|156398335|ref|XP_001638144.1|predicted protein [Nematostella vectensis] 2e-149; gi|1036029214|gb|ANJ59783.1|alpha carbonic anhydrase 7 [Exaiptasia pallida] 3e-127; gi|999984030|gb|KXJ23171.1|Carbonic anhydrase 7 [Exaiptasia pallida] 6e-127; gi|1036029172|gb|ANJ59762.1|alpha carbonic anhydrase 1 [Stylophora pistillata] 1e-94; gi|169402112|gb|ACA53457.1|alpha carbonic anhydrase [Stylophora pistillata] 7e-56; gi|403310239|embl|CCJ09594.1|carbonic anyhdrase, partial [Patella vulgata] 1e-52; gi|308321240|gb|ADO27772.1|carbonic anhydrase [Ictalurus furcatus] 6e-52; gi|1083295622|ref|XP_018596813.1|PREDICTED: carbonic anhydrase-like [Scleropages formosus] 8e-52; gi|319068359|ref|NP_001187560.1|carbonic anhydrase [Ictalurus punctatus] 1e-51; | ||||||
| NvERTx.4.37378 | R-8 | 0.997054170012 | E-5 | 0.999999895906 | Q4R8Y5 | IQ domain-containing protein D | gi|999972710|gb|KXJ11884.1|IQ domain-containing protein D [Exaiptasia pallida], 1e-41 |
| other nr_hits | gi|156395185|ref|XP_001636992.1|predicted protein [Nematostella vectensis] 5e-34; gi|1005450327|ref|XP_015762362.1|PREDICTED: IQ domain-containing protein D-like [Acropora digitifera] 1e-31; gi|291224212|ref|XP_002732097.1|PREDICTED: IQ domain-containing protein D-like [Saccoglossus kowalevskii] 3e-16; gi|390357293|ref|XP_784878.3|PREDICTED: IQ domain-containing protein D [Strongylocentrotus purpuratus] 1e-11; gi|919089441|ref|XP_013381737.1|PREDICTED: IQ domain-containing protein D-like [Lingula anatina] 1e-09; gi|676466568|ref|XP_009057608.1|hypothetical protein LOTGIDRAFT_83213, partial [Lottia gigantea] 1e-09; gi|762126431|ref|XP_011446272.1|PREDICTED: IQ domain-containing protein D [Crassostrea gigas] 3e-09; gi|405968992|gb|EKC34008.1|IQ domain-containing protein D [Crassostrea gigas] 3e-09; gi|871205275|ref|XP_012934688.1|PREDICTED: IQ domain-containing protein D-like [Aplysia californica] 4e-09; | ||||||
| NvERTx.4.37379 | R-8 | 0.997054170012 | E-5 | 0.999999895906 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.176388 | R-8 | 0.997054170012 | E-5 | 0.999999895906 | Q17QH9 | IQ domain-containing protein D | gi|999972710|gb|KXJ11884.1|IQ domain-containing protein D [Exaiptasia pallida], 1e-131 |
| other nr_hits | gi|1005450327|ref|XP_015762362.1|PREDICTED: IQ domain-containing protein D-like [Acropora digitifera] 2e-97; gi|156395185|ref|XP_001636992.1|predicted protein [Nematostella vectensis] 6e-85; gi|291224212|ref|XP_002732097.1|PREDICTED: IQ domain-containing protein D-like [Saccoglossus kowalevskii] 6e-79; gi|390357293|ref|XP_784878.3|PREDICTED: IQ domain-containing protein D [Strongylocentrotus purpuratus] 4e-73; gi|260799724|ref|XP_002594834.1|hypothetical protein BRAFLDRAFT_124432 [Branchiostoma floridae] 2e-70; gi|1126159230|ref|XP_019618705.1|PREDICTED: IQ domain-containing protein D-like [Branchiostoma belcheri] 7e-70; gi|676466568|ref|XP_009057608.1|hypothetical protein LOTGIDRAFT_83213, partial [Lottia gigantea] 3e-66; gi|762126431|ref|XP_011446272.1|PREDICTED: IQ domain-containing protein D [Crassostrea gigas] 3e-65; gi|919089441|ref|XP_013381737.1|PREDICTED: IQ domain-containing protein D-like [Lingula anatina] 7e-64; | ||||||
| NvERTx.4.137929 | R-7 | 0.999850264884 | E-5 | 0.999999895803 | Q4RGM4 | 39S ribosomal protein L33, mitochondrial | gi|156400092|ref|XP_001638834.1|predicted protein [Nematostella vectensis], 1e-21 |
| other nr_hits | gi|909143505|gb|KNE67751.1|ribosomal protein L33 [Allomyces macrogynus ATCC 38327] 8e-13; gi|576988028|gb|EUC60752.1|39S ribosomal protein L33 [Rhizoctonia solani AG-3 Rhs1AP] 9e-12; gi|639561915|gb|KDN39633.1|hypothetical protein RSAG8_08661, partial [Rhizoctonia solani AG-8 WAC10335] 1e-11; gi|301109667|ref|XP_002903914.1|conserved hypothetical protein [Phytophthora infestans T30-4] 2e-11; gi|82252455|sp|RM33_TETNG|Q4RGM4.1|RecName: Full=39S ribosomal protein L33, mitochondrial; Short=L33mt; Short=MRP-L33; Flags: Precursor >CAG12458.1 unnamed protein product [Tetraodon nigroviridis] 3e-11; gi|635372372|embl|CCI39593.1|unnamed protein product [Albugo candida] 3e-11; gi|669147634|ref|XP_008609379.1|50S ribosomal protein L33 [Saprolegnia diclina VS20] 3e-11; gi|1026951089|ref|XP_016612760.1|ribosomal protein L33 [Spizellomyces punctatus DAOM BR117] 4e-11; gi|1072252322|ref|XP_018411109.1|PREDICTED: 39S ribosomal protein L33, mitochondrial [Nanorana parkeri] 6e-11; | ||||||
| NvERTx.4.137930 | R-7 | 0.999850264884 | E-5 | 0.999999895803 | Q4RGM4 | 39S ribosomal protein L33, mitochondrial | gi|156400092|ref|XP_001638834.1|predicted protein [Nematostella vectensis], 1e-31 |
| other nr_hits | gi|909143505|gb|KNE67751.1|ribosomal protein L33 [Allomyces macrogynus ATCC 38327] 5e-11; gi|1071052600|ref|XP_018275000.1|hypothetical protein CC85DRAFT_331549 [Cutaneotrichosporon oleaginosus] 4e-10; gi|1027071780|ref|XP_016639272.1|hypothetical protein SAPIO_CDS9339 [Scedosporium apiospermum] 2e-09; gi|576988028|gb|EUC60752.1|39S ribosomal protein L33 [Rhizoctonia solani AG-3 Rhs1AP] 2e-09; gi|676380449|ref|XP_009033503.1|hypothetical protein AURANDRAFT_20043 [Aureococcus anophagefferens] 2e-09; gi|1072252322|ref|XP_018411109.1|PREDICTED: 39S ribosomal protein L33, mitochondrial [Nanorana parkeri] 3e-09; gi|1026951089|ref|XP_016612760.1|ribosomal protein L33 [Spizellomyces punctatus DAOM BR117] 3e-09; gi|639561915|gb|KDN39633.1|hypothetical protein RSAG8_08661, partial [Rhizoctonia solani AG-8 WAC10335] 3e-09; gi|955480766|ref|XP_014653711.1|serine carboxypeptidase [Moesziomyces antarcticus] 6e-09; | ||||||
| NvERTx.4.113932 | R-3 | 0.959481386901 | E-5 | 0.999999895064 | Q8HZ57 | Coiled-coil alpha-helical rod protein 1 | gi|919078350|ref|XP_013420328.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X1 [Lingula anatina], 1e-76 |
| other nr_hits | gi|919078352|ref|XP_013420329.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X2 [Lingula anatina] 1e-76; gi|1126173647|ref|XP_019618967.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like [Branchiostoma belcheri] 5e-62; gi|1069381382|ref|XP_018085659.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like [Xenopus laevis] 8e-62; gi|999966501|gb|KXJ05675.1|Coiled-coil alpha-helical rod protein 1, partial [Exaiptasia pallida] 1e-60; gi|1062920801|ref|XP_017951636.1|PREDICTED: coiled-coil alpha-helical rod protein 1 isoform X1 [Xenopus tropicalis] 7e-60; gi|183986659|ref|NP_001116918.1|coiled-coil alpha-helical rod protein 1 [Xenopus tropicalis] 7e-60; gi|847153146|ref|XP_012823433.1|PREDICTED: coiled-coil alpha-helical rod protein 1 isoform X2 [Xenopus tropicalis] 3e-59; gi|1083287768|ref|XP_018592566.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X1 [Scleropages formosus] 9e-56; gi|1083287770|ref|XP_018592567.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X2 [Scleropages formosus] 9e-56; | ||||||
| NvERTx.4.113933 | R-3 | 0.959481386901 | E-5 | 0.999999895064 | Q8HZ57 | Coiled-coil alpha-helical rod protein 1 | gi|919078350|ref|XP_013420328.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X1 [Lingula anatina], 6e-78 |
| other nr_hits | gi|919078352|ref|XP_013420329.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X2 [Lingula anatina] 6e-78; gi|1126173647|ref|XP_019618967.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like [Branchiostoma belcheri] 1e-63; gi|1069381382|ref|XP_018085659.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like [Xenopus laevis] 2e-63; gi|999966501|gb|KXJ05675.1|Coiled-coil alpha-helical rod protein 1, partial [Exaiptasia pallida] 2e-62; gi|1062920801|ref|XP_017951636.1|PREDICTED: coiled-coil alpha-helical rod protein 1 isoform X1 [Xenopus tropicalis] 2e-61; gi|183986659|ref|NP_001116918.1|coiled-coil alpha-helical rod protein 1 [Xenopus tropicalis] 2e-61; gi|847153146|ref|XP_012823433.1|PREDICTED: coiled-coil alpha-helical rod protein 1 isoform X2 [Xenopus tropicalis] 7e-61; gi|1083287768|ref|XP_018592566.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X1 [Scleropages formosus] 3e-57; gi|1083287770|ref|XP_018592567.1|PREDICTED: coiled-coil alpha-helical rod protein 1-like isoform X2 [Scleropages formosus] 3e-57; | ||||||
| NvERTx.4.220296 | R-3 | 0.959481386901 | E-5 | 0.999999895064 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.5895 | R-5 | 0.995255308474 | E-5 | 0.999999893883 | No_Uniprotmatch | No_Description | gi|156361830|ref|XP_001625487.1|predicted protein [Nematostella vectensis], 7e-21 |
| other nr_hits | NA | ||||||
| NvERTx.4.70342 | R-5 | 0.995255308474 | E-5 | 0.999999893883 | A0A0L0BSP9_LUCCU | Uncharacterized protein {ECO:0000313|EMBL:KNC23067.1} | gi|156361830|ref|XP_001625487.1|predicted protein [Nematostella vectensis], 2e-150 |
| other nr_hits | gi|1005472399|ref|XP_015773066.1|PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC107351287 [Acropora digitifera] 1e-28; gi|1005461519|ref|XP_015767841.1|PREDICTED: uncharacterized protein LOC107346548 [Acropora digitifera] 9e-16; gi|906460271|gb|KNC23067.1|hypothetical protein FF38_01498 [Lucilia cuprina] 2e-11; gi|985424751|ref|XP_015377876.1|PREDICTED: uncharacterized protein LOC107172115 [Diuraphis noxia] 2e-10; gi|1080039252|ref|XP_018580150.1|PREDICTED: uncharacterized protein LOC108917849 [Anoplophora glabripennis] 9e-10; gi|1005446692|ref|XP_015760635.1|PREDICTED: uncharacterized protein LOC107339815 [Acropora digitifera] 5e-09; gi|195997389|ref|XP_002108563.1|hypothetical protein TRIADDRAFT_51596 [Trichoplax adhaerens] 5e-09; gi|1101334862|ref|XP_018903545.1|PREDICTED: uncharacterized protein LOC109034712 [Bemisia tabaci] 3e-08; gi|939669883|ref|XP_014281774.1|PREDICTED: uncharacterized protein LOC106684298 [Halyomorpha halys] 4e-08; | ||||||
| NvERTx.4.14874 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.133960 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | P23128 | Putative ATP-dependent RNA helicase me31b | gi|156380897|ref|XP_001632003.1|predicted protein [Nematostella vectensis], 9e-149 |
| other nr_hits | gi|449678669|ref|XP_002164714.2|PREDICTED: probable ATP-dependent RNA helicase DDX6 [Hydra vulgaris] 2e-120; gi|999975868|gb|KXJ15042.1|putative ATP-dependent RNA helicase me31b [Exaiptasia pallida] 2e-119; gi|545917330|gb|JAB57347.1|putative translation initiation factor 4f helicase subunit eif-4a [Corethrella appendiculata] 2e-113; gi|1005421097|ref|XP_015760109.1|PREDICTED: probable ATP-dependent RNA helicase ddx6 [Acropora digitifera] 3e-113; gi|443697450|gb|ELT97926.1|hypothetical protein CAPTEDRAFT_155246 [Capitella teleta] 4e-113; gi|1078809963|ref|XP_018496669.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Galendromus occidentalis] 4e-112; gi|157118999|ref|XP_001659287.1|AAEL008500-PA [Aedes aegypti] 4e-112; gi|1131317293|gb|JAV09006.1|putative translation initiation factor 4f helicase subunit eif-4a, partial [Nyssomyia neivai] 5e-112; gi|1131317723|gb|JAV09221.1|putative translation initiation factor 4f helicase subunit eif-4a [Nyssomyia neivai] 5e-112; | ||||||
| NvERTx.4.133961 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | P23128 | Putative ATP-dependent RNA helicase me31b | gi|156380897|ref|XP_001632003.1|predicted protein [Nematostella vectensis], 2e-240 |
| other nr_hits | gi|449678669|ref|XP_002164714.2|PREDICTED: probable ATP-dependent RNA helicase DDX6 [Hydra vulgaris] 2e-198; gi|999975868|gb|KXJ15042.1|putative ATP-dependent RNA helicase me31b [Exaiptasia pallida] 1e-194; gi|443697450|gb|ELT97926.1|hypothetical protein CAPTEDRAFT_155246 [Capitella teleta] 4e-190; gi|1078809963|ref|XP_018496669.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Galendromus occidentalis] 1e-188; gi|1005421097|ref|XP_015760109.1|PREDICTED: probable ATP-dependent RNA helicase ddx6 [Acropora digitifera] 3e-188; gi|675365809|gb|KFM58711.1|putative ATP-dependent RNA helicase me31b, partial [Stegodyphus mimosarum] 3e-187; gi|926607370|ref|XP_013772166.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Limulus polyphemus] 4e-187; gi|1126217372|ref|XP_019642737.1|PREDICTED: probable ATP-dependent RNA helicase DDX6 [Branchiostoma belcheri] 3e-186; gi|762094796|ref|XP_011429888.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Crassostrea gigas] 5e-186; | ||||||
| NvERTx.4.133962 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | P23128 | Putative ATP-dependent RNA helicase me31b | gi|156380897|ref|XP_001632003.1|predicted protein [Nematostella vectensis], 2e-240 |
| other nr_hits | gi|449678669|ref|XP_002164714.2|PREDICTED: probable ATP-dependent RNA helicase DDX6 [Hydra vulgaris] 2e-198; gi|999975868|gb|KXJ15042.1|putative ATP-dependent RNA helicase me31b [Exaiptasia pallida] 1e-194; gi|443697450|gb|ELT97926.1|hypothetical protein CAPTEDRAFT_155246 [Capitella teleta] 4e-190; gi|1078809963|ref|XP_018496669.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Galendromus occidentalis] 1e-188; gi|1005421097|ref|XP_015760109.1|PREDICTED: probable ATP-dependent RNA helicase ddx6 [Acropora digitifera] 3e-188; gi|675365809|gb|KFM58711.1|putative ATP-dependent RNA helicase me31b, partial [Stegodyphus mimosarum] 3e-187; gi|926607370|ref|XP_013772166.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Limulus polyphemus] 4e-187; gi|1126217372|ref|XP_019642737.1|PREDICTED: probable ATP-dependent RNA helicase DDX6 [Branchiostoma belcheri] 3e-186; gi|762094796|ref|XP_011429888.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Crassostrea gigas] 5e-186; | ||||||
| NvERTx.4.133963 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | P23128 | Putative ATP-dependent RNA helicase me31b | gi|156380897|ref|XP_001632003.1|predicted protein [Nematostella vectensis], 9e-149 |
| other nr_hits | gi|449678669|ref|XP_002164714.2|PREDICTED: probable ATP-dependent RNA helicase DDX6 [Hydra vulgaris] 2e-120; gi|999975868|gb|KXJ15042.1|putative ATP-dependent RNA helicase me31b [Exaiptasia pallida] 2e-119; gi|545917330|gb|JAB57347.1|putative translation initiation factor 4f helicase subunit eif-4a [Corethrella appendiculata] 2e-113; gi|1005421097|ref|XP_015760109.1|PREDICTED: probable ATP-dependent RNA helicase ddx6 [Acropora digitifera] 3e-113; gi|443697450|gb|ELT97926.1|hypothetical protein CAPTEDRAFT_155246 [Capitella teleta] 4e-113; gi|1078809963|ref|XP_018496669.1|PREDICTED: putative ATP-dependent RNA helicase me31b [Galendromus occidentalis] 4e-112; gi|157118999|ref|XP_001659287.1|AAEL008500-PA [Aedes aegypti] 4e-112; gi|1131317293|gb|JAV09006.1|putative translation initiation factor 4f helicase subunit eif-4a, partial [Nyssomyia neivai] 5e-112; gi|1131317723|gb|JAV09221.1|putative translation initiation factor 4f helicase subunit eif-4a [Nyssomyia neivai] 5e-112; | ||||||
| NvERTx.4.139241 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.139242 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.139243 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.139244 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.139245 | R-7 | 0.999903855383 | E-5 | 0.999999893201 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.77149 | R-2 | 0.751864985964 | E-5 | 0.999999892846 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.77151 | R-2 | 0.751864985964 | E-5 | 0.999999892846 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.77156 | R-2 | 0.751864985964 | E-5 | 0.999999892846 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.77157 | R-2 | 0.751864985964 | E-5 | 0.999999892846 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.218030 | R-2 | 0.751864985964 | E-5 | 0.999999892846 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.93987 | R-7 | 0.611870629739 | E-5 | 0.999999891226 | No_Uniprotmatch | No_Description | gi|156342135|ref|XP_001620888.1|hypothetical protein NEMVEDRAFT_v1g222602 [Nematostella vectensis], 8e-30 |
| other nr_hits | NA | ||||||
| NvERTx.4.93988 | R-7 | 0.611870629739 | E-5 | 0.999999891226 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.93989 | R-7 | 0.611870629739 | E-5 | 0.999999891226 | No_Uniprotmatch | No_Description | gi|156342135|ref|XP_001620888.1|hypothetical protein NEMVEDRAFT_v1g222602 [Nematostella vectensis], 6e-41 |
| other nr_hits | gi|1154807074|ref|WP_078722831.1|hypothetical protein [Streptococcus pneumoniae] 3e-05; | ||||||
| NvERTx.4.93990 | R-7 | 0.611870629739 | E-5 | 0.999999891226 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.130796 | R-6 | 0.98388975189 | E-5 | 0.999999889078 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.130804 | R-6 | 0.98388975189 | E-5 | 0.999999889078 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.130805 | R-6 | 0.98388975189 | E-5 | 0.999999889078 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.6254 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.41878 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.170324 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 6e-146 |
| other nr_hits | gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 7e-84; gi|1126167255|ref|XP_019615481.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1-like [Branchiostoma belcheri] 1e-66; gi|919031635|ref|XP_013399441.1|PREDICTED: transcription elongation regulator 1-like isoform X1 [Lingula anatina] 1e-65; gi|919031637|ref|XP_013399442.1|PREDICTED: transcription elongation regulator 1-like isoform X2 [Lingula anatina] 1e-65; gi|585660653|ref|XP_006816425.1|PREDICTED: transcription elongation regulator 1-like [Saccoglossus kowalevskii] 9e-65; gi|926626052|ref|XP_013778552.1|PREDICTED: transcription elongation regulator 1-like [Limulus polyphemus] 2e-64; gi|972976412|ref|XP_015204794.1|PREDICTED: transcription elongation regulator 1 [Lepisosteus oculatus] 3e-64; gi|1062853229|ref|XP_017947688.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Xenopus tropicalis] 5e-64; gi|301611603|ref|XP_002935314.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Xenopus tropicalis] 5e-64; | ||||||
| NvERTx.4.170325 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 2e-107 |
| other nr_hits | gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 1e-67; gi|1126167255|ref|XP_019615481.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1-like [Branchiostoma belcheri] 9e-56; gi|1041107368|ref|XP_017276904.1|PREDICTED: transcription elongation regulator 1 isoform X3 [Kryptolebias marmoratus] 1e-55; gi|1041107364|ref|XP_017276902.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Kryptolebias marmoratus] 1e-55; gi|1041107366|ref|XP_017276903.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Kryptolebias marmoratus] 1e-55; gi|1041107370|ref|XP_017276906.1|PREDICTED: transcription elongation regulator 1 isoform X4 [Kryptolebias marmoratus] 1e-55; gi|1041107372|ref|XP_017276907.1|PREDICTED: transcription elongation regulator 1 isoform X5 [Kryptolebias marmoratus] 1e-55; gi|1041107374|ref|XP_017276908.1|PREDICTED: transcription elongation regulator 1 isoform X6 [Kryptolebias marmoratus] 1e-55; gi|961894953|ref|XP_014901510.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Poecilia latipinna] 3e-55; | ||||||
| NvERTx.4.170326 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | Q8CGF7 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 3e-30 |
| other nr_hits | gi|999983939|gb|KXJ23080.1|Transcription elongation regulator 1 [Exaiptasia pallida] 9e-13; gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 1e-10; gi|743727736|ref|XP_010957911.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1 [Camelus bactrianus] 5e-09; gi|625259695|ref|XP_007621268.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Cricetulus griseus] 5e-09; gi|632948415|ref|XP_007889586.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Callorhinchus milii] 8e-09; gi|632948417|ref|XP_007889587.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Callorhinchus milii] 8e-09; gi|632948419|ref|XP_007889588.1|PREDICTED: transcription elongation regulator 1 isoform X3 [Callorhinchus milii] 8e-09; gi|632948421|ref|XP_007889590.1|PREDICTED: transcription elongation regulator 1 isoform X4 [Callorhinchus milii] 8e-09; gi|632948423|ref|XP_007889591.1|PREDICTED: transcription elongation regulator 1 isoform X5 [Callorhinchus milii] 8e-09; | ||||||
| NvERTx.4.170327 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 5e-252 |
| other nr_hits | gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 1e-129; gi|1126167255|ref|XP_019615481.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1-like [Branchiostoma belcheri] 5e-105; gi|556977973|ref|XP_005996147.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Latimeria chalumnae] 2e-104; gi|556977976|ref|XP_005996148.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Latimeria chalumnae] 2e-104; gi|1109010923|ref|XP_010751997.2|PREDICTED: transcription elongation regulator 1 isoform X1 [Larimichthys crocea] 2e-104; gi|1109010927|ref|XP_010751999.2|PREDICTED: transcription elongation regulator 1 isoform X3 [Larimichthys crocea] 2e-104; gi|999983939|gb|KXJ23080.1|Transcription elongation regulator 1 [Exaiptasia pallida] 5e-104; gi|1109010925|ref|XP_010751998.2|PREDICTED: transcription elongation regulator 1 isoform X2 [Larimichthys crocea] 2e-103; gi|1109010929|ref|XP_010752000.2|PREDICTED: transcription elongation regulator 1 isoform X4 [Larimichthys crocea] 2e-103; | ||||||
| NvERTx.4.170328 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 8e-143 |
| other nr_hits | gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 2e-82; gi|1126167255|ref|XP_019615481.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1-like [Branchiostoma belcheri] 2e-66; gi|919031635|ref|XP_013399441.1|PREDICTED: transcription elongation regulator 1-like isoform X1 [Lingula anatina] 2e-65; gi|919031637|ref|XP_013399442.1|PREDICTED: transcription elongation regulator 1-like isoform X2 [Lingula anatina] 2e-65; gi|972976412|ref|XP_015204794.1|PREDICTED: transcription elongation regulator 1 [Lepisosteus oculatus] 2e-64; gi|585660653|ref|XP_006816425.1|PREDICTED: transcription elongation regulator 1-like [Saccoglossus kowalevskii] 2e-63; gi|556977973|ref|XP_005996147.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Latimeria chalumnae] 2e-63; gi|556977976|ref|XP_005996148.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Latimeria chalumnae] 2e-63; gi|1062853229|ref|XP_017947688.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Xenopus tropicalis] 2e-63; | ||||||
| NvERTx.4.170329 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 6e-146 |
| other nr_hits | gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 7e-84; gi|1126167255|ref|XP_019615481.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1-like [Branchiostoma belcheri] 1e-66; gi|919031635|ref|XP_013399441.1|PREDICTED: transcription elongation regulator 1-like isoform X1 [Lingula anatina] 1e-65; gi|919031637|ref|XP_013399442.1|PREDICTED: transcription elongation regulator 1-like isoform X2 [Lingula anatina] 1e-65; gi|585660653|ref|XP_006816425.1|PREDICTED: transcription elongation regulator 1-like [Saccoglossus kowalevskii] 9e-65; gi|926626052|ref|XP_013778552.1|PREDICTED: transcription elongation regulator 1-like [Limulus polyphemus] 2e-64; gi|972976412|ref|XP_015204794.1|PREDICTED: transcription elongation regulator 1 [Lepisosteus oculatus] 3e-64; gi|1062853229|ref|XP_017947688.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Xenopus tropicalis] 5e-64; gi|301611603|ref|XP_002935314.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Xenopus tropicalis] 5e-64; | ||||||
| NvERTx.4.170330 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 4e-209 |
| other nr_hits | gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 1e-99; gi|1109010923|ref|XP_010751997.2|PREDICTED: transcription elongation regulator 1 isoform X1 [Larimichthys crocea] 2e-80; gi|1109010927|ref|XP_010751999.2|PREDICTED: transcription elongation regulator 1 isoform X3 [Larimichthys crocea] 2e-80; gi|1126167255|ref|XP_019615481.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1-like [Branchiostoma belcheri] 4e-80; gi|1109010925|ref|XP_010751998.2|PREDICTED: transcription elongation regulator 1 isoform X2 [Larimichthys crocea] 1e-79; gi|1109010929|ref|XP_010752000.2|PREDICTED: transcription elongation regulator 1 isoform X4 [Larimichthys crocea] 1e-79; gi|808859570|gb|KKF11676.1|Transcription elongation regulator 1-like protein, partial [Larimichthys crocea] 1e-79; gi|1158875135|ref|XP_020341542.1|transcription elongation regulator 1-like isoform X2 [Oncorhynchus kisutch] 2e-79; gi|657585695|ref|XP_008297056.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Stegastes partitus] 2e-79; | ||||||
| NvERTx.4.170331 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 4e-209 |
| other nr_hits | gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 1e-99; gi|1109010923|ref|XP_010751997.2|PREDICTED: transcription elongation regulator 1 isoform X1 [Larimichthys crocea] 2e-80; gi|1109010927|ref|XP_010751999.2|PREDICTED: transcription elongation regulator 1 isoform X3 [Larimichthys crocea] 2e-80; gi|1126167255|ref|XP_019615481.1|PREDICTED: LOW QUALITY PROTEIN: transcription elongation regulator 1-like [Branchiostoma belcheri] 4e-80; gi|1109010925|ref|XP_010751998.2|PREDICTED: transcription elongation regulator 1 isoform X2 [Larimichthys crocea] 1e-79; gi|1109010929|ref|XP_010752000.2|PREDICTED: transcription elongation regulator 1 isoform X4 [Larimichthys crocea] 1e-79; gi|808859570|gb|KKF11676.1|Transcription elongation regulator 1-like protein, partial [Larimichthys crocea] 1e-79; gi|1158875135|ref|XP_020341542.1|transcription elongation regulator 1-like isoform X2 [Oncorhynchus kisutch] 2e-79; gi|657585695|ref|XP_008297056.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Stegastes partitus] 2e-79; | ||||||
| NvERTx.4.170332 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | O14776 | Transcription elongation regulator 1 | gi|156390268|ref|XP_001635193.1|predicted protein [Nematostella vectensis], 6e-91 |
| other nr_hits | gi|999983939|gb|KXJ23080.1|Transcription elongation regulator 1 [Exaiptasia pallida] 3e-57; gi|1005424704|ref|XP_015777364.1|PREDICTED: transcription elongation regulator 1-like [Acropora digitifera] 7e-54; gi|1041107368|ref|XP_017276904.1|PREDICTED: transcription elongation regulator 1 isoform X3 [Kryptolebias marmoratus] 3e-49; gi|1041107364|ref|XP_017276902.1|PREDICTED: transcription elongation regulator 1 isoform X1 [Kryptolebias marmoratus] 3e-49; gi|1041107366|ref|XP_017276903.1|PREDICTED: transcription elongation regulator 1 isoform X2 [Kryptolebias marmoratus] 3e-49; gi|1041107370|ref|XP_017276906.1|PREDICTED: transcription elongation regulator 1 isoform X4 [Kryptolebias marmoratus] 3e-49; gi|1041107372|ref|XP_017276907.1|PREDICTED: transcription elongation regulator 1 isoform X5 [Kryptolebias marmoratus] 3e-49; gi|1041107374|ref|XP_017276908.1|PREDICTED: transcription elongation regulator 1 isoform X6 [Kryptolebias marmoratus] 3e-49; gi|1109010925|ref|XP_010751998.2|PREDICTED: transcription elongation regulator 1 isoform X2 [Larimichthys crocea] 5e-49; | ||||||
| NvERTx.4.215751 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.229346 | R-7 | 0.904741324736 | E-5 | 0.999999886614 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.105419 | R-1 | 0.939300266028 | E-5 | 0.999999884033 | Q96GM5 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 | gi|156398524|ref|XP_001638238.1|predicted protein [Nematostella vectensis], 4e-224 |
| other nr_hits | gi|999989195|gb|KXJ28237.1|SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 [Exaiptasia pallida] 5e-212; gi|260783506|ref|XP_002586815.1|hypothetical protein BRAFLDRAFT_247762 [Branchiostoma floridae] 4e-173; gi|676438489|ref|XP_009048546.1|hypothetical protein LOTGIDRAFT_140600 [Lottia gigantea] 8e-172; gi|632987185|ref|XP_007910652.1|PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 [Callorhinchus milii] 1e-171; gi|38198635|ref|NP_938172.1|SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 [Danio rerio] 2e-170; gi|410919329|ref|XP_003973137.1|PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 [Takifugu rubripes] 2e-170; gi|47220711|embl|CAG11780.1|unnamed protein product [Tetraodon nigroviridis] 3e-170; gi|583974938|ref|XP_006782849.1|PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1-like [Neolamprologus brichardi] 3e-170; gi|554866589|ref|XP_005943316.1|PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 [Haplochromis burtoni] 4e-170; | ||||||
