Explore Mfuzz Clusters
Mfuzz cluster
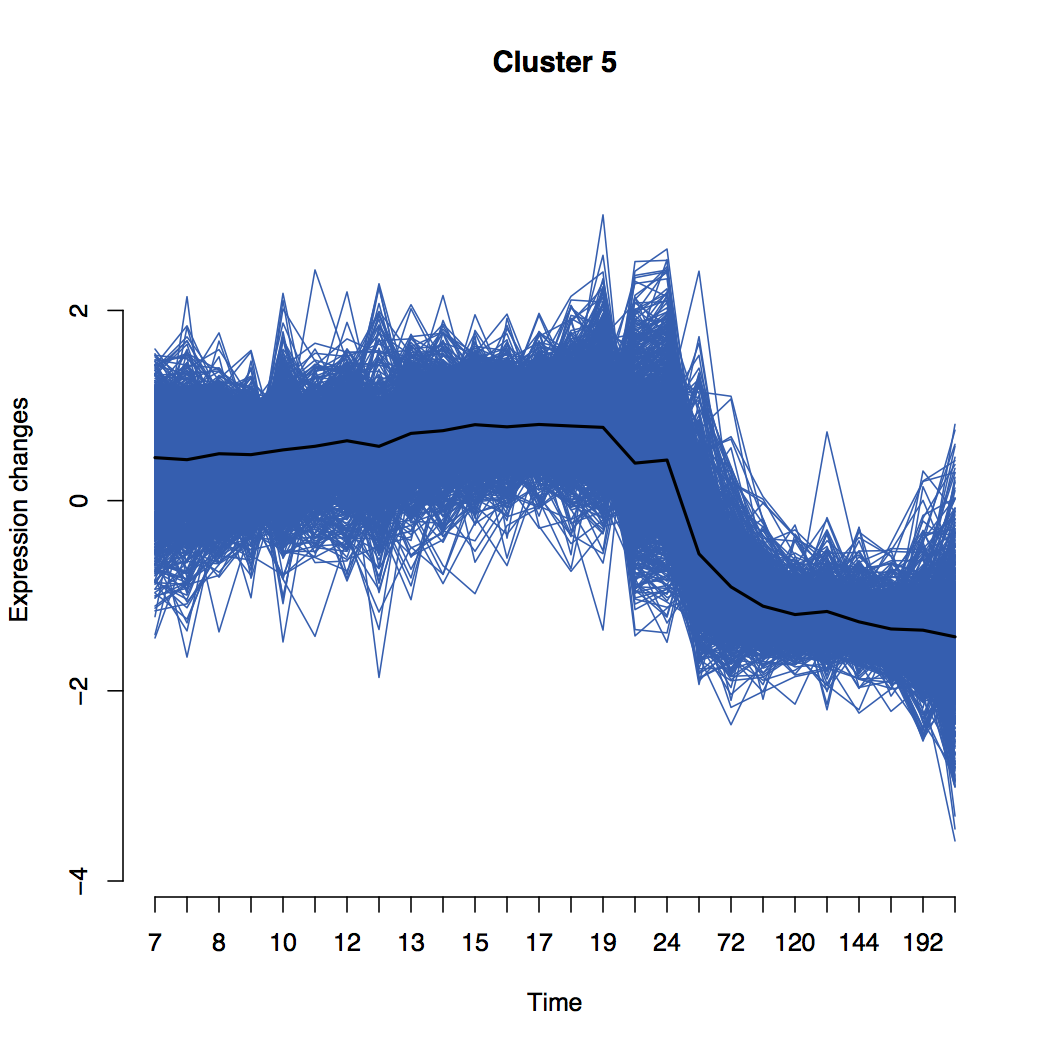
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.135200 | R-3 | 0.564621034925 | E-5 | 0.999999941079 | No_Uniprotmatch | No_Description | gi|156344522|ref|XP_001621216.1|hypothetical protein NEMVEDRAFT_v1g222238 [Nematostella vectensis], 5e-71 |
| other nr_hits | gi|156346844|ref|XP_001621548.1|hypothetical protein NEMVEDRAFT_v1g221855 [Nematostella vectensis] 5e-70; gi|156337874|ref|XP_001619907.1|hypothetical protein NEMVEDRAFT_v1g223698 [Nematostella vectensis] 5e-68; gi|156346841|ref|XP_001621547.1|hypothetical protein NEMVEDRAFT_v1g221857 [Nematostella vectensis] 5e-68; gi|156346918|ref|XP_001621575.1|hypothetical protein NEMVEDRAFT_v1g221821 [Nematostella vectensis] 5e-68; gi|156348638|ref|XP_001621924.1|hypothetical protein NEMVEDRAFT_v1g221407 [Nematostella vectensis] 4e-54; gi|156351535|ref|XP_001622555.1|predicted protein [Nematostella vectensis] 6e-54; gi|156340909|ref|XP_001620591.1|hypothetical protein NEMVEDRAFT_v1g222941 [Nematostella vectensis] 4e-52; gi|156339492|ref|XP_001620177.1|hypothetical protein NEMVEDRAFT_v1g223370 [Nematostella vectensis] 8e-45; gi|156346920|ref|XP_001621576.1|hypothetical protein NEMVEDRAFT_v1g221822 [Nematostella vectensis] 8e-45; | ||||||
| NvERTx.4.135204 | R-3 | 0.564621034925 | E-5 | 0.999999941079 | No_Uniprotmatch | No_Description | gi|156340109|ref|XP_001620355.1|hypothetical protein NEMVEDRAFT_v1g223199 [Nematostella vectensis], 3e-34 |
| other nr_hits | gi|156344522|ref|XP_001621216.1|hypothetical protein NEMVEDRAFT_v1g222238 [Nematostella vectensis] 3e-34; gi|156346844|ref|XP_001621548.1|hypothetical protein NEMVEDRAFT_v1g221855 [Nematostella vectensis] 7e-34; gi|156340909|ref|XP_001620591.1|hypothetical protein NEMVEDRAFT_v1g222941 [Nematostella vectensis] 1e-33; gi|156337874|ref|XP_001619907.1|hypothetical protein NEMVEDRAFT_v1g223698 [Nematostella vectensis] 7e-32; gi|156346841|ref|XP_001621547.1|hypothetical protein NEMVEDRAFT_v1g221857 [Nematostella vectensis] 7e-32; gi|156346918|ref|XP_001621575.1|hypothetical protein NEMVEDRAFT_v1g221821 [Nematostella vectensis] 3e-31; gi|156351535|ref|XP_001622555.1|predicted protein [Nematostella vectensis] 7e-24; gi|156348638|ref|XP_001621924.1|hypothetical protein NEMVEDRAFT_v1g221407 [Nematostella vectensis] 1e-20; gi|156340570|ref|XP_001620488.1|hypothetical protein NEMVEDRAFT_v1g223061 [Nematostella vectensis] 1e-18; | ||||||
| NvERTx.4.135205 | R-3 | 0.564621034925 | E-5 | 0.999999941079 | No_Uniprotmatch | No_Description | gi|156344522|ref|XP_001621216.1|hypothetical protein NEMVEDRAFT_v1g222238 [Nematostella vectensis], 5e-71 |
| other nr_hits | gi|156346844|ref|XP_001621548.1|hypothetical protein NEMVEDRAFT_v1g221855 [Nematostella vectensis] 5e-70; gi|156337874|ref|XP_001619907.1|hypothetical protein NEMVEDRAFT_v1g223698 [Nematostella vectensis] 5e-68; gi|156346841|ref|XP_001621547.1|hypothetical protein NEMVEDRAFT_v1g221857 [Nematostella vectensis] 5e-68; gi|156346918|ref|XP_001621575.1|hypothetical protein NEMVEDRAFT_v1g221821 [Nematostella vectensis] 5e-68; gi|156348638|ref|XP_001621924.1|hypothetical protein NEMVEDRAFT_v1g221407 [Nematostella vectensis] 4e-54; gi|156351535|ref|XP_001622555.1|predicted protein [Nematostella vectensis] 6e-54; gi|156340909|ref|XP_001620591.1|hypothetical protein NEMVEDRAFT_v1g222941 [Nematostella vectensis] 4e-52; gi|156339492|ref|XP_001620177.1|hypothetical protein NEMVEDRAFT_v1g223370 [Nematostella vectensis] 8e-45; gi|156346920|ref|XP_001621576.1|hypothetical protein NEMVEDRAFT_v1g221822 [Nematostella vectensis] 8e-45; | ||||||
| NvERTx.4.110966 | R-8 | 0.999391678503 | E-5 | 0.999999940891 | Q08CH6 | Ciliogenesis-associated TTC17-interacting protein | gi|999989008|gb|KXJ28072.1|Ciliogenesis-associated TTC17-interacting protein [Exaiptasia pallida], 4e-114 |
| other nr_hits | gi|291228944|ref|XP_002734427.1|PREDICTED: uncharacterized protein C2orf62 homolog isoform X1 [Saccoglossus kowalevskii] 5e-84; gi|156361297|ref|XP_001625454.1|predicted protein [Nematostella vectensis] 7e-83; gi|585658740|ref|XP_006816064.1|PREDICTED: uncharacterized protein C2orf62 homolog isoform X2 [Saccoglossus kowalevskii] 2e-82; gi|919007558|ref|XP_013388667.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like [Lingula anatina] 2e-81; gi|1126185620|ref|XP_019625486.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like [Branchiostoma belcheri] 1e-78; gi|762156651|ref|XP_011416459.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein [Crassostrea gigas] 6e-77; gi|390339969|ref|XP_793449.3|PREDICTED: ciliogenesis-associated TTC17-interacting protein [Strongylocentrotus purpuratus] 5e-76; gi|871263248|ref|XP_012943597.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like [Aplysia californica] 3e-74; gi|908460605|ref|XP_013085271.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like, partial [Biomphalaria glabrata] 2e-73; | ||||||
| NvERTx.4.110961 | R-8 | 0.999391678503 | E-5 | 0.999999940891 | Q08CH6 | Ciliogenesis-associated TTC17-interacting protein | gi|999989008|gb|KXJ28072.1|Ciliogenesis-associated TTC17-interacting protein [Exaiptasia pallida], 2e-108 |
| other nr_hits | gi|156361297|ref|XP_001625454.1|predicted protein [Nematostella vectensis] 7e-83; gi|291228944|ref|XP_002734427.1|PREDICTED: uncharacterized protein C2orf62 homolog isoform X1 [Saccoglossus kowalevskii] 1e-81; gi|585658740|ref|XP_006816064.1|PREDICTED: uncharacterized protein C2orf62 homolog isoform X2 [Saccoglossus kowalevskii] 3e-81; gi|919007558|ref|XP_013388667.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like [Lingula anatina] 3e-78; gi|1126185620|ref|XP_019625486.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like [Branchiostoma belcheri] 3e-76; gi|762156651|ref|XP_011416459.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein [Crassostrea gigas] 1e-75; gi|390339969|ref|XP_793449.3|PREDICTED: ciliogenesis-associated TTC17-interacting protein [Strongylocentrotus purpuratus] 6e-74; gi|908460605|ref|XP_013085271.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like, partial [Biomphalaria glabrata] 2e-72; gi|871263248|ref|XP_012943597.1|PREDICTED: ciliogenesis-associated TTC17-interacting protein-like [Aplysia californica] 2e-72; | ||||||
| NvERTx.4.211399 | R-6 | 0.981416109627 | E-5 | 0.99999994056 | No_Uniprotmatch | No_Description | gi|156354150|ref|XP_001623264.1|predicted protein [Nematostella vectensis], 2e-08 |
| other nr_hits | NA | ||||||
| NvERTx.4.69995 | R-5 | 0.999973665794 | E-5 | 0.999999938915 | Q14674 | Separin | gi|156391219|ref|XP_001635666.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999989170|gb|KXJ28212.1|Separin [Exaiptasia pallida] 2e-205; gi|780154110|ref|XP_011682126.1|PREDICTED: separin [Strongylocentrotus purpuratus] 1e-92; gi|919021941|ref|XP_013394828.1|PREDICTED: separin-like [Lingula anatina] 2e-92; gi|919057291|ref|XP_013410481.1|PREDICTED: separin-like [Lingula anatina] 2e-91; gi|1126205985|ref|XP_019636551.1|PREDICTED: LOW QUALITY PROTEIN: separin-like [Branchiostoma belcheri] 2e-90; gi|957847509|ref|XP_014674242.1|PREDICTED: uncharacterized protein LOC106814439 [Priapulus caudatus] 2e-90; gi|762082372|ref|XP_011423994.1|PREDICTED: uncharacterized protein LOC105325911 [Crassostrea gigas] 2e-90; gi|828211608|ref|XP_012558716.1|PREDICTED: separin-like [Hydra vulgaris] 6e-88; gi|961085852|ref|XP_014770196.1|PREDICTED: uncharacterized protein LOC106869135 [Octopus bimaculoides] 7e-88; | ||||||
| NvERTx.4.69996 | R-5 | 0.999973665794 | E-5 | 0.999999938915 | Q14674 | Separin | gi|156391219|ref|XP_001635666.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999989170|gb|KXJ28212.1|Separin [Exaiptasia pallida] 4e-308; gi|780154110|ref|XP_011682126.1|PREDICTED: separin [Strongylocentrotus purpuratus] 2e-92; gi|919021941|ref|XP_013394828.1|PREDICTED: separin-like [Lingula anatina] 3e-92; gi|919057291|ref|XP_013410481.1|PREDICTED: separin-like [Lingula anatina] 3e-91; gi|1126205985|ref|XP_019636551.1|PREDICTED: LOW QUALITY PROTEIN: separin-like [Branchiostoma belcheri] 4e-90; gi|957847509|ref|XP_014674242.1|PREDICTED: uncharacterized protein LOC106814439 [Priapulus caudatus] 4e-90; gi|762082372|ref|XP_011423994.1|PREDICTED: uncharacterized protein LOC105325911 [Crassostrea gigas] 4e-90; gi|828211608|ref|XP_012558716.1|PREDICTED: separin-like [Hydra vulgaris] 1e-87; gi|961085852|ref|XP_014770196.1|PREDICTED: uncharacterized protein LOC106869135 [Octopus bimaculoides] 1e-87; | ||||||
| NvERTx.4.69997 | R-5 | 0.999973665794 | E-5 | 0.999999938915 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.209389 | R-5 | 0.999973665794 | E-5 | 0.999999938915 | P60330 | Separin | gi|156391219|ref|XP_001635666.1|predicted protein [Nematostella vectensis], 5e-32 |
| other nr_hits | gi|999989170|gb|KXJ28212.1|Separin [Exaiptasia pallida] 3e-19; gi|971447119|ref|XP_015128533.1|PREDICTED: separin isoform X1 [Gallus gallus] 7e-15; gi|971447121|ref|XP_015128534.1|PREDICTED: separin isoform X2 [Gallus gallus] 7e-15; gi|1121841571|ref|XP_019467152.1|PREDICTED: separin [Meleagris gallopavo] 2e-14; gi|946698588|ref|XP_014437021.1|PREDICTED: LOW QUALITY PROTEIN: separin [Pelodiscus sinensis] 2e-14; gi|1004005069|ref|XP_015705841.1|PREDICTED: separin isoform X1 [Coturnix japonica] 2e-14; gi|1004005072|ref|XP_015705843.1|PREDICTED: separin isoform X2 [Coturnix japonica] 2e-14; gi|919021941|ref|XP_013394828.1|PREDICTED: separin-like [Lingula anatina] 3e-14; gi|696969436|ref|XP_009555107.1|PREDICTED: separin [Cuculus canorus] 3e-14; | ||||||
| NvERTx.4.219461 | R-5 | 0.999973665794 | E-5 | 0.999999938915 | No_Uniprotmatch | No_Description | gi|156391219|ref|XP_001635666.1|predicted protein [Nematostella vectensis], 6e-14 |
| other nr_hits | NA | ||||||
| NvERTx.4.58236 | R-6 | 0.999912953832 | E-5 | 0.999999937018 | Q9D967 | Magnesium-dependent phosphatase 1 | gi|156350481|ref|XP_001622301.1|predicted protein [Nematostella vectensis], 4e-94 |
| other nr_hits | gi|511867898|ref|XP_004755293.1|PREDICTED: magnesium-dependent phosphatase 1 [Mustela putorius furo] 5e-49; gi|410961990|ref|XP_003987561.1|PREDICTED: magnesium-dependent phosphatase 1 isoform X2 [Felis catus] 2e-48; gi|836707503|ref|XP_004609729.2|PREDICTED: magnesium-dependent phosphatase 1 [Sorex araneus] 3e-48; gi|1111218279|ref|XP_019284887.1|PREDICTED: magnesium-dependent phosphatase 1 [Panthera pardus] 4e-48; gi|926627866|ref|XP_013779526.1|PREDICTED: magnesium-dependent phosphatase 1-like [Limulus polyphemus] 4e-48; gi|557024445|ref|XP_006012405.1|PREDICTED: magnesium-dependent phosphatase 1 [Latimeria chalumnae] 1e-47; gi|472365082|ref|XP_004402099.1|PREDICTED: magnesium-dependent phosphatase 1 [Odobenus rosmarus divergens] 2e-47; gi|533123562|ref|XP_005377078.1|PREDICTED: magnesium-dependent phosphatase 1 isoform X1 [Chinchilla lanigera] 3e-47; gi|157823059|ref|NP_001099509.1|magnesium-dependent phosphatase 1 [Rattus norvegicus] 4e-47; | ||||||
| NvERTx.4.58237 | R-6 | 0.999912953832 | E-5 | 0.999999937018 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.58238 | R-6 | 0.999912953832 | E-5 | 0.999999937018 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.113983 | R-5 | 0.77388862949 | E-5 | 0.99999993675 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.113984 | R-5 | 0.77388862949 | E-5 | 0.99999993675 | Q96SK2 | Transmembrane protein 209 | gi|156393577|ref|XP_001636404.1|predicted protein [Nematostella vectensis], 3e-72 |
| other nr_hits | gi|1005469002|ref|XP_015771421.1|PREDICTED: transmembrane protein 209-like isoform X1 [Acropora digitifera] 3e-57; gi|999971379|gb|KXJ10553.1|Transmembrane protein 209 [Exaiptasia pallida] 1e-42; gi|957838731|ref|XP_014669607.1|PREDICTED: transmembrane protein 209-like [Priapulus caudatus] 3e-42; gi|961069297|ref|XP_014771592.1|PREDICTED: transmembrane protein 209-like [Octopus bimaculoides] 1e-39; gi|926628682|ref|XP_013779960.1|PREDICTED: transmembrane protein 209-like [Limulus polyphemus] 1e-39; gi|918338861|gb|KOF99950.1|hypothetical protein OCBIM_22009527mg, partial [Octopus bimaculoides] 1e-39; gi|908479831|ref|XP_013091590.1|PREDICTED: transmembrane protein 209-like [Biomphalaria glabrata] 1e-39; gi|909793585|ref|XP_013153025.1|PREDICTED: transmembrane protein 209 isoform X1 [Falco peregrinus] 2e-39; gi|828213035|ref|XP_004209062.2|PREDICTED: transmembrane protein 209-like, partial [Hydra vulgaris] 2e-39; | ||||||
| NvERTx.4.113986 | R-5 | 0.77388862949 | E-5 | 0.99999993675 | Q96SK2 | Transmembrane protein 209 | gi|156393577|ref|XP_001636404.1|predicted protein [Nematostella vectensis], 3e-72 |
| other nr_hits | gi|1005469002|ref|XP_015771421.1|PREDICTED: transmembrane protein 209-like isoform X1 [Acropora digitifera] 3e-57; gi|999971379|gb|KXJ10553.1|Transmembrane protein 209 [Exaiptasia pallida] 1e-42; gi|957838731|ref|XP_014669607.1|PREDICTED: transmembrane protein 209-like [Priapulus caudatus] 3e-42; gi|961069297|ref|XP_014771592.1|PREDICTED: transmembrane protein 209-like [Octopus bimaculoides] 1e-39; gi|926628682|ref|XP_013779960.1|PREDICTED: transmembrane protein 209-like [Limulus polyphemus] 1e-39; gi|918338861|gb|KOF99950.1|hypothetical protein OCBIM_22009527mg, partial [Octopus bimaculoides] 1e-39; gi|908479831|ref|XP_013091590.1|PREDICTED: transmembrane protein 209-like [Biomphalaria glabrata] 1e-39; gi|909793585|ref|XP_013153025.1|PREDICTED: transmembrane protein 209 isoform X1 [Falco peregrinus] 2e-39; gi|828213035|ref|XP_004209062.2|PREDICTED: transmembrane protein 209-like, partial [Hydra vulgaris] 2e-39; | ||||||
| NvERTx.4.113987 | R-5 | 0.77388862949 | E-5 | 0.99999993675 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.113988 | R-5 | 0.77388862949 | E-5 | 0.99999993675 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.113989 | R-5 | 0.77388862949 | E-5 | 0.99999993675 | Q8BRG8 | Transmembrane protein 209 | gi|156393577|ref|XP_001636404.1|predicted protein [Nematostella vectensis], 6e-140 |
| other nr_hits | gi|1005469002|ref|XP_015771421.1|PREDICTED: transmembrane protein 209-like isoform X1 [Acropora digitifera] 3e-125; gi|919097196|ref|XP_013385276.1|PREDICTED: transmembrane protein 209-like [Lingula anatina] 5e-105; gi|919072093|ref|XP_013417334.1|PREDICTED: transmembrane protein 209-like [Lingula anatina] 7e-105; gi|1005469006|ref|XP_015771423.1|PREDICTED: transmembrane protein 209-like isoform X3 [Acropora digitifera] 4e-102; gi|1126201657|ref|XP_019634182.1|PREDICTED: transmembrane protein 209-like [Branchiostoma belcheri] 2e-100; gi|291221607|ref|XP_002730812.1|PREDICTED: transmembrane protein 209-like [Saccoglossus kowalevskii] 4e-100; gi|694840351|ref|XP_009460865.1|PREDICTED: transmembrane protein 209 [Nipponia nippon] 3e-90; gi|690460681|ref|XP_009329291.1|PREDICTED: transmembrane protein 209 [Pygoscelis adeliae] 3e-90; gi|677532603|gb|KFQ95064.1|Transmembrane protein 209, partial [Nipponia nippon] 3e-90; | ||||||
| NvERTx.4.52076 | R-1 | 0.56135837792 | E-5 | 0.999999936743 | Q8VC31 | Coiled-coil domain-containing protein 9 | gi|156370096|ref|XP_001628308.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999966949|gb|KXJ06123.1|Coiled-coil domain-containing protein 9 [Exaiptasia pallida] 7e-57; gi|1005448692|ref|XP_015761601.1|PREDICTED: coiled-coil domain-containing protein 9-like [Acropora digitifera] 4e-36; gi|1008788586|ref|XP_015865242.1|PREDICTED: coiled-coil domain-containing protein 9 isoform X8 [Peromyscus maniculatus bairdii] 8e-24; gi|1126189342|ref|XP_019627529.1|PREDICTED: coiled-coil domain-containing protein 9-like isoform X3 [Branchiostoma belcheri] 2e-22; gi|1126189338|ref|XP_019627527.1|PREDICTED: coiled-coil domain-containing protein 9-like isoform X1 [Branchiostoma belcheri] 2e-22; gi|1126189340|ref|XP_019627528.1|PREDICTED: coiled-coil domain-containing protein 9-like isoform X2 [Branchiostoma belcheri] 4e-22; gi|831498377|ref|XP_012709711.1|PREDICTED: coiled-coil domain-containing protein 9 [Fundulus heteroclitus] 1e-21; gi|974111331|ref|XP_015254877.1|PREDICTED: coiled-coil domain-containing protein 9 isoform X2 [Cyprinodon variegatus] 5e-20; gi|584021219|ref|XP_006805286.1|PREDICTED: coiled-coil domain-containing protein 9-like [Neolamprologus brichardi] 5e-20; | ||||||
| NvERTx.4.52077 | R-1 | 0.56135837792 | E-5 | 0.999999936743 | Q8VC31 | Coiled-coil domain-containing protein 9 | gi|156370096|ref|XP_001628308.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999966949|gb|KXJ06123.1|Coiled-coil domain-containing protein 9 [Exaiptasia pallida] 7e-57; gi|1005448692|ref|XP_015761601.1|PREDICTED: coiled-coil domain-containing protein 9-like [Acropora digitifera] 4e-36; gi|1008788586|ref|XP_015865242.1|PREDICTED: coiled-coil domain-containing protein 9 isoform X8 [Peromyscus maniculatus bairdii] 7e-24; gi|1126189342|ref|XP_019627529.1|PREDICTED: coiled-coil domain-containing protein 9-like isoform X3 [Branchiostoma belcheri] 2e-22; gi|1126189338|ref|XP_019627527.1|PREDICTED: coiled-coil domain-containing protein 9-like isoform X1 [Branchiostoma belcheri] 2e-22; gi|1126189340|ref|XP_019627528.1|PREDICTED: coiled-coil domain-containing protein 9-like isoform X2 [Branchiostoma belcheri] 4e-22; gi|831498377|ref|XP_012709711.1|PREDICTED: coiled-coil domain-containing protein 9 [Fundulus heteroclitus] 1e-21; gi|974111331|ref|XP_015254877.1|PREDICTED: coiled-coil domain-containing protein 9 isoform X2 [Cyprinodon variegatus] 5e-20; gi|584021219|ref|XP_006805286.1|PREDICTED: coiled-coil domain-containing protein 9-like [Neolamprologus brichardi] 5e-20; | ||||||
| NvERTx.4.207315 | R-1 | 0.56135837792 | E-5 | 0.999999936743 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.39480 | R-8 | 0.23662063175 | E-5 | 0.999999936662 | P53367 | Arfaptin-1 | gi|156408447|ref|XP_001641868.1|predicted protein [Nematostella vectensis], 3e-112 |
| other nr_hits | gi|999973027|gb|KXJ12201.1|Arfaptin-1 [Exaiptasia pallida] 7e-97; gi|1035268069|ref|XP_016887540.1|PREDICTED: arfaptin-2-like isoform X1 [Cynoglossus semilaevis] 4e-73; gi|884955017|ref|XP_012990069.1|PREDICTED: arfaptin-2 isoform X1 [Esox lucius] 1e-72; gi|884955020|ref|XP_012990070.1|PREDICTED: arfaptin-2 isoform X2 [Esox lucius] 1e-72; gi|884955024|ref|XP_012990072.1|PREDICTED: arfaptin-2 isoform X4 [Esox lucius] 1e-72; gi|584010450|ref|XP_006800157.1|PREDICTED: arfaptin-2-like isoform X1 [Neolamprologus brichardi] 2e-72; gi|584010454|ref|XP_006800159.1|PREDICTED: arfaptin-2-like isoform X3 [Neolamprologus brichardi] 2e-72; gi|584010456|ref|XP_006800160.1|PREDICTED: arfaptin-2-like isoform X4 [Neolamprologus brichardi] 2e-72; gi|1110883317|ref|XP_019219527.1|PREDICTED: arfaptin-2 isoform X6 [Oreochromis niloticus] 2e-72; | ||||||
| NvERTx.4.39481 | R-8 | 0.23662063175 | E-5 | 0.999999936662 | P53367 | Arfaptin-1 | gi|999973027|gb|KXJ12201.1|Arfaptin-1 [Exaiptasia pallida], 5e-120 |
| other nr_hits | gi|156408447|ref|XP_001641868.1|predicted protein [Nematostella vectensis] 3e-112; gi|1005419546|ref|XP_015753228.1|PREDICTED: arfaptin-2-like [Acropora digitifera] 2e-78; gi|736309501|ref|XP_010795565.1|PREDICTED: arfaptin-2-like isoform X2 [Notothenia coriiceps] 7e-73; gi|884955017|ref|XP_012990069.1|PREDICTED: arfaptin-2 isoform X1 [Esox lucius] 2e-72; gi|884955020|ref|XP_012990070.1|PREDICTED: arfaptin-2 isoform X2 [Esox lucius] 2e-72; gi|884955024|ref|XP_012990072.1|PREDICTED: arfaptin-2 isoform X4 [Esox lucius] 2e-72; gi|736309497|ref|XP_010795564.1|PREDICTED: arfaptin-2-like isoform X1 [Notothenia coriiceps] 2e-72; gi|736309510|ref|XP_010795567.1|PREDICTED: arfaptin-2-like isoform X4 [Notothenia coriiceps] 2e-72; gi|584010450|ref|XP_006800157.1|PREDICTED: arfaptin-2-like isoform X1 [Neolamprologus brichardi] 2e-72; | ||||||
| NvERTx.4.7076 | R-5 | 0.878409749373 | E-5 | 0.999999936437 | NA | NA | None |
| other nr_hits | NA | ||||||
| NvERTx.4.55996 | R-5 | 0.878409749373 | E-5 | 0.999999936437 | Q90YG6 | Transcription initiation factor IIA subunit 2 | gi|156402941|ref|XP_001639848.1|predicted protein [Nematostella vectensis], 2e-39 |
| other nr_hits | gi|999988032|gb|KXJ27135.1|Transcription initiation factor IIA subunit 2 [Exaiptasia pallida] 1e-36; gi|1005444839|ref|XP_015759756.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Acropora digitifera] 3e-34; gi|291222339|ref|XP_002731169.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Saccoglossus kowalevskii] 4e-34; gi|828200982|ref|XP_012555098.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Hydra vulgaris] 5e-33; gi|919058400|ref|XP_013411084.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Lingula anatina] 1e-31; gi|1129984310|ref|XP_019748825.1|PREDICTED: transcription initiation factor IIA subunit 2 [Hippocampus comes] 2e-31; gi|1049273453|ref|XP_017577076.1|PREDICTED: transcription initiation factor IIA subunit 2 [Pygocentrus nattereri] 2e-31; gi|1042251675|ref|XP_017332841.1|PREDICTED: transcription initiation factor IIA subunit 2 [Ictalurus punctatus] 2e-31; gi|1007736083|ref|XP_015808679.1|PREDICTED: transcription initiation factor IIA subunit 2 [Nothobranchius furzeri] 2e-31; | ||||||
| NvERTx.4.55997 | R-5 | 0.878409749373 | E-5 | 0.999999936437 | Q90YG6 | Transcription initiation factor IIA subunit 2 | gi|156402941|ref|XP_001639848.1|predicted protein [Nematostella vectensis], 2e-39 |
| other nr_hits | gi|999988032|gb|KXJ27135.1|Transcription initiation factor IIA subunit 2 [Exaiptasia pallida] 1e-36; gi|1005444839|ref|XP_015759756.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Acropora digitifera] 3e-34; gi|291222339|ref|XP_002731169.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Saccoglossus kowalevskii] 4e-34; gi|828200982|ref|XP_012555098.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Hydra vulgaris] 5e-33; gi|919058400|ref|XP_013411084.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Lingula anatina] 1e-31; gi|1129984310|ref|XP_019748825.1|PREDICTED: transcription initiation factor IIA subunit 2 [Hippocampus comes] 2e-31; gi|1049273453|ref|XP_017577076.1|PREDICTED: transcription initiation factor IIA subunit 2 [Pygocentrus nattereri] 2e-31; gi|1042251675|ref|XP_017332841.1|PREDICTED: transcription initiation factor IIA subunit 2 [Ictalurus punctatus] 2e-31; gi|1007736083|ref|XP_015808679.1|PREDICTED: transcription initiation factor IIA subunit 2 [Nothobranchius furzeri] 2e-31; | ||||||
| NvERTx.4.55998 | R-5 | 0.878409749373 | E-5 | 0.999999936437 | Q90YG6 | Transcription initiation factor IIA subunit 2 | gi|156402941|ref|XP_001639848.1|predicted protein [Nematostella vectensis], 8e-19 |
| other nr_hits | gi|999988032|gb|KXJ27135.1|Transcription initiation factor IIA subunit 2 [Exaiptasia pallida] 5e-16; gi|1005444839|ref|XP_015759756.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Acropora digitifera] 5e-14; gi|919058400|ref|XP_013411084.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Lingula anatina] 1e-13; gi|780034756|ref|XP_011667230.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Strongylocentrotus purpuratus] 1e-13; gi|291222339|ref|XP_002731169.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Saccoglossus kowalevskii] 1e-13; gi|313231289|embl|CBY08404.1|unnamed protein product [Oikopleura dioica] 2e-13; gi|676441805|ref|XP_009049607.1|hypothetical protein LOTGIDRAFT_238741 [Lottia gigantea] 3e-13; gi|443692949|gb|ELT94433.1|hypothetical protein CAPTEDRAFT_146781 [Capitella teleta] 7e-13; gi|961104873|ref|XP_014776786.1|PREDICTED: transcription initiation factor IIA subunit 2-like [Octopus bimaculoides] 9e-13; | ||||||
| NvERTx.4.220377 | R-5 | 0.878409749373 | E-5 | 0.999999936437 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.69984 | R-7 | 0.999947180911 | E-5 | 0.999999935802 | Q92620 | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 | gi|1005456486|ref|XP_015765374.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16-like [Acropora digitifera], 0e+00 |
| other nr_hits | gi|926611828|ref|XP_013794146.1|PREDICTED: LOW QUALITY PROTEIN: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16-like [Limulus polyphemus] 0e+00; gi|831551814|ref|XP_012728481.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Fundulus heteroclitus] 0e+00; gi|974083594|ref|XP_015239698.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X1 [Cyprinodon variegatus] 0e+00; gi|974083598|ref|XP_015239700.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X2 [Cyprinodon variegatus] 0e+00; gi|1025469603|ref|XP_016536386.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X1 [Poecilia formosa] 0e+00; gi|961846571|ref|XP_014888770.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X1 [Poecilia latipinna] 0e+00; gi|658842322|ref|XP_008403553.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X1 [Poecilia reticulata] 0e+00; gi|658842324|ref|XP_008403554.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X2 [Poecilia reticulata] 0e+00; gi|617456593|ref|XP_007570505.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X2 [Poecilia formosa] 0e+00; | ||||||
| NvERTx.4.69985 | R-7 | 0.999947180911 | E-5 | 0.999999935802 | Q17R09 | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 | gi|1005456486|ref|XP_015765374.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16-like [Acropora digitifera], 0e+00 |
| other nr_hits | gi|926611828|ref|XP_013794146.1|PREDICTED: LOW QUALITY PROTEIN: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16-like [Limulus polyphemus] 0e+00; gi|942166822|ref|XP_014346344.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Latimeria chalumnae] 0e+00; gi|1143399946|ref|XP_019954372.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X2 [Paralichthys olivaceus] 0e+00; gi|640807821|ref|XP_008060416.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Carlito syrichta] 0e+00; gi|1143399944|ref|XP_019954371.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X1 [Paralichthys olivaceus] 0e+00; gi|471412558|ref|XP_004387828.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Trichechus manatus latirostris] 0e+00; gi|831551814|ref|XP_012728481.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Fundulus heteroclitus] 0e+00; gi|831278787|ref|XP_012670336.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Clupea harengus] 0e+00; gi|634846733|ref|XP_007938509.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Orycteropus afer afer] 0e+00; | ||||||
| NvERTx.4.69986 | R-7 | 0.999947180911 | E-5 | 0.999999935802 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.69987 | R-7 | 0.999947180911 | E-5 | 0.999999935802 | Q17R09 | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 | gi|1005456486|ref|XP_015765374.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16-like [Acropora digitifera], 0e+00 |
| other nr_hits | gi|942166822|ref|XP_014346344.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Latimeria chalumnae] 0e+00; gi|926611828|ref|XP_013794146.1|PREDICTED: LOW QUALITY PROTEIN: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16-like [Limulus polyphemus] 0e+00; gi|831551814|ref|XP_012728481.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Fundulus heteroclitus] 0e+00; gi|41053698|ref|NP_957170.1|pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Danio rerio] 0e+00; gi|1143399946|ref|XP_019954372.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X2 [Paralichthys olivaceus] 0e+00; gi|1143399944|ref|XP_019954371.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X1 [Paralichthys olivaceus] 0e+00; gi|1042254772|ref|XP_017333585.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [Ictalurus punctatus] 0e+00; gi|974083594|ref|XP_015239698.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X1 [Cyprinodon variegatus] 0e+00; gi|974083598|ref|XP_015239700.1|PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 isoform X2 [Cyprinodon variegatus] 0e+00; | ||||||
| NvERTx.4.119518 | R-7 | 0.999799737677 | E-5 | 0.999999935117 | Q7ZTY4 | Histone-binding protein RBBP7 | gi|156386844|ref|XP_001634121.1|predicted protein [Nematostella vectensis], 3e-248 |
| other nr_hits | gi|999987011|gb|KXJ26136.1|Histone-binding protein RBBP7 [Exaiptasia pallida] 2e-237; gi|1022772871|gb|KZS17028.1|Histone-binding protein Caf1 [Daphnia magna] 4e-229; gi|321465178|gb|EFX76181.1|hypothetical protein DAPPUDRAFT_306287 [Daphnia pulex] 4e-229; gi|755980664|ref|XP_011309322.1|PREDICTED: probable histone-binding protein Caf1 [Fopius arisanus] 6e-229; gi|805824644|ref|XP_012152261.1|PREDICTED: probable histone-binding protein Caf1 isoform X3 [Megachile rotundata] 7e-229; gi|1070182191|ref|XP_018349438.1|PREDICTED: probable histone-binding protein Caf1 isoform X2 [Trachymyrmex septentrionalis] 1e-228; gi|1069676550|ref|XP_018300542.1|PREDICTED: probable histone-binding protein Caf1 isoform X2 [Trachymyrmex zeteki] 1e-228; gi|1068411665|ref|XP_018052090.1|PREDICTED: probable histone-binding protein Caf1 isoform X5 [Atta colombica] 1e-228; gi|746857633|ref|XP_011059557.1|PREDICTED: probable histone-binding protein Caf1 isoform X4 [Acromyrmex echinatior] 1e-228; | ||||||
| NvERTx.4.173691 | R-7 | 0.999799737677 | E-5 | 0.999999935117 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.173696 | R-7 | 0.999799737677 | E-5 | 0.999999935117 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.227656 | R-7 | 0.999799737677 | E-5 | 0.999999935117 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.6695 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.7873 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.138611 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | Q6ZQ88 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 5e-41 |
| other nr_hits | gi|999976294|gb|KXJ15468.1|Lysine-specific histone demethylase 1A [Exaiptasia pallida] 7e-39; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 5e-32; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 5e-32; gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 1e-24; gi|961069191|ref|XP_014771266.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Octopus bimaculoides] 3e-23; gi|1101645626|ref|XP_018922034.1|PREDICTED: lysine-specific histone demethylase 1A [Cyprinus carpio] 6e-23; gi|966643293|gb|KTF78205.1|hypothetical protein cypCar_00014188 [Cyprinus carpio] 6e-23; gi|528506410|ref|XP_005158840.1|PREDICTED: lysine-specific histone demethylase 1A isoform X3 [Danio rerio] 6e-23; gi|966673679|gb|KTG07060.1|hypothetical protein cypCar_00029020 [Cyprinus carpio] 8e-23; | ||||||
| NvERTx.4.138620 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | O60341 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 7e-107 |
| other nr_hits | gi|1005437378|ref|XP_015756129.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Acropora digitifera] 2e-81; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 2e-81; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 2e-81; gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 2e-81; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 1e-79; gi|1126190580|ref|XP_019628210.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Branchiostoma belcheri] 9e-76; gi|1126190594|ref|XP_019628217.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X10 [Branchiostoma belcheri] 9e-76; gi|1126190584|ref|XP_019628212.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X5 [Branchiostoma belcheri] 9e-76; gi|260791152|ref|XP_002590604.1|hypothetical protein BRAFLDRAFT_123611 [Branchiostoma floridae] 6e-75; | ||||||
| NvERTx.4.138621 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | O60341 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 2e-111 |
| other nr_hits | gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 3e-89; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 3e-89; gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 6e-82; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 2e-73; gi|961069191|ref|XP_014771266.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Octopus bimaculoides] 4e-73; gi|528506410|ref|XP_005158840.1|PREDICTED: lysine-specific histone demethylase 1A isoform X3 [Danio rerio] 1e-72; gi|966643293|gb|KTF78205.1|hypothetical protein cypCar_00014188 [Cyprinus carpio] 2e-72; gi|966673679|gb|KTG07060.1|hypothetical protein cypCar_00029020 [Cyprinus carpio] 2e-72; gi|1025286685|ref|XP_016372099.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Sinocyclocheilus rhinocerous] 4e-72; | ||||||
| NvERTx.4.138612 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | O60341 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 9e-84 |
| other nr_hits | gi|1005437378|ref|XP_015756129.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Acropora digitifera] 3e-67; gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 3e-67; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 3e-67; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 6e-64; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 1e-63; gi|1019015755|ref|XP_016082284.1|PREDICTED: lysine-specific histone demethylase 1A [Ornithorhynchus anatinus] 6e-56; gi|47230008|embl|CAG10422.1|unnamed protein product, partial [Tetraodon nigroviridis] 2e-54; gi|632954842|ref|XP_007893176.1|PREDICTED: lysine-specific histone demethylase 1A, partial [Callorhinchus milii] 1e-53; gi|1083268022|ref|XP_018588686.1|PREDICTED: lysine-specific histone demethylase 1A isoform X1 [Scleropages formosus] 1e-52; | ||||||
| NvERTx.4.138613 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | O60341 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 1e-84 |
| other nr_hits | gi|1005437378|ref|XP_015756129.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Acropora digitifera] 2e-68; gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 2e-68; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 2e-68; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 6e-65; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 1e-64; gi|1019015755|ref|XP_016082284.1|PREDICTED: lysine-specific histone demethylase 1A [Ornithorhynchus anatinus] 9e-56; gi|632954842|ref|XP_007893176.1|PREDICTED: lysine-specific histone demethylase 1A, partial [Callorhinchus milii] 1e-54; gi|47230008|embl|CAG10422.1|unnamed protein product, partial [Tetraodon nigroviridis] 3e-54; gi|1083268022|ref|XP_018588686.1|PREDICTED: lysine-specific histone demethylase 1A isoform X1 [Scleropages formosus] 1e-53; | ||||||
| NvERTx.4.138615 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | O60341 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 0e+00; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 0e+00; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 0e+00; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 1e-309; gi|1005437378|ref|XP_015756129.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Acropora digitifera] 4e-304; gi|1126190584|ref|XP_019628212.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X5 [Branchiostoma belcheri] 2e-273; gi|1126190594|ref|XP_019628217.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X10 [Branchiostoma belcheri] 8e-273; gi|1126190578|ref|XP_019628209.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Branchiostoma belcheri] 3e-272; gi|1126190580|ref|XP_019628210.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Branchiostoma belcheri] 4e-272; | ||||||
| NvERTx.4.138616 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.138617 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | Q6ZQ88 | Lysine-specific histone demethylase 1A | gi|1005437378|ref|XP_015756129.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Acropora digitifera], 8e-211 |
| other nr_hits | gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 8e-211; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 8e-211; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 8e-211; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 9e-210; gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis] 4e-209; gi|1126190584|ref|XP_019628212.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X5 [Branchiostoma belcheri] 2e-184; gi|1126190594|ref|XP_019628217.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X10 [Branchiostoma belcheri] 2e-184; gi|1126190578|ref|XP_019628209.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Branchiostoma belcheri] 4e-183; gi|1126190580|ref|XP_019628210.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Branchiostoma belcheri] 6e-183; | ||||||
| NvERTx.4.138618 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | Q6ZQ88 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 0e+00; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 0e+00; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 0e+00; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 1e-308; gi|1005437378|ref|XP_015756129.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Acropora digitifera] 6e-303; gi|1126190584|ref|XP_019628212.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X5 [Branchiostoma belcheri] 2e-272; gi|1126190594|ref|XP_019628217.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X10 [Branchiostoma belcheri] 7e-272; gi|1126190578|ref|XP_019628209.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Branchiostoma belcheri] 4e-271; gi|1126190592|ref|XP_019628216.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X9 [Branchiostoma belcheri] 4e-271; | ||||||
| NvERTx.4.138619 | R-7 | 0.525172696055 | E-5 | 0.999999934025 | O60341 | Lysine-specific histone demethylase 1A | gi|156383987|ref|XP_001633113.1|predicted protein [Nematostella vectensis], 6e-18 |
| other nr_hits | gi|1005437378|ref|XP_015756129.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X4 [Acropora digitifera] 4e-09; gi|1005437374|ref|XP_015756127.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X2 [Acropora digitifera] 4e-09; gi|1005437372|ref|XP_015756126.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X1 [Acropora digitifera] 4e-09; gi|1005437376|ref|XP_015756128.1|PREDICTED: lysine-specific histone demethylase 1A-like isoform X3 [Acropora digitifera] 4e-09; gi|356467211|gb|AET09736.1|hypothetical protein p3_17, partial [Acropora millepora] 8e-09; gi|1101645626|ref|XP_018922034.1|PREDICTED: lysine-specific histone demethylase 1A [Cyprinus carpio] 3e-07; gi|902929938|ref|XP_013053091.1|PREDICTED: lysine-specific histone demethylase 1A [Anser cygnoides domesticus] 3e-07; gi|831318959|ref|XP_012692249.1|PREDICTED: lysine-specific histone demethylase 1A isoform X2 [Clupea harengus] 3e-07; gi|831318957|ref|XP_012692248.1|PREDICTED: lysine-specific histone demethylase 1A isoform X1 [Clupea harengus] 3e-07; | ||||||
