Explore Mfuzz Clusters
Mfuzz cluster
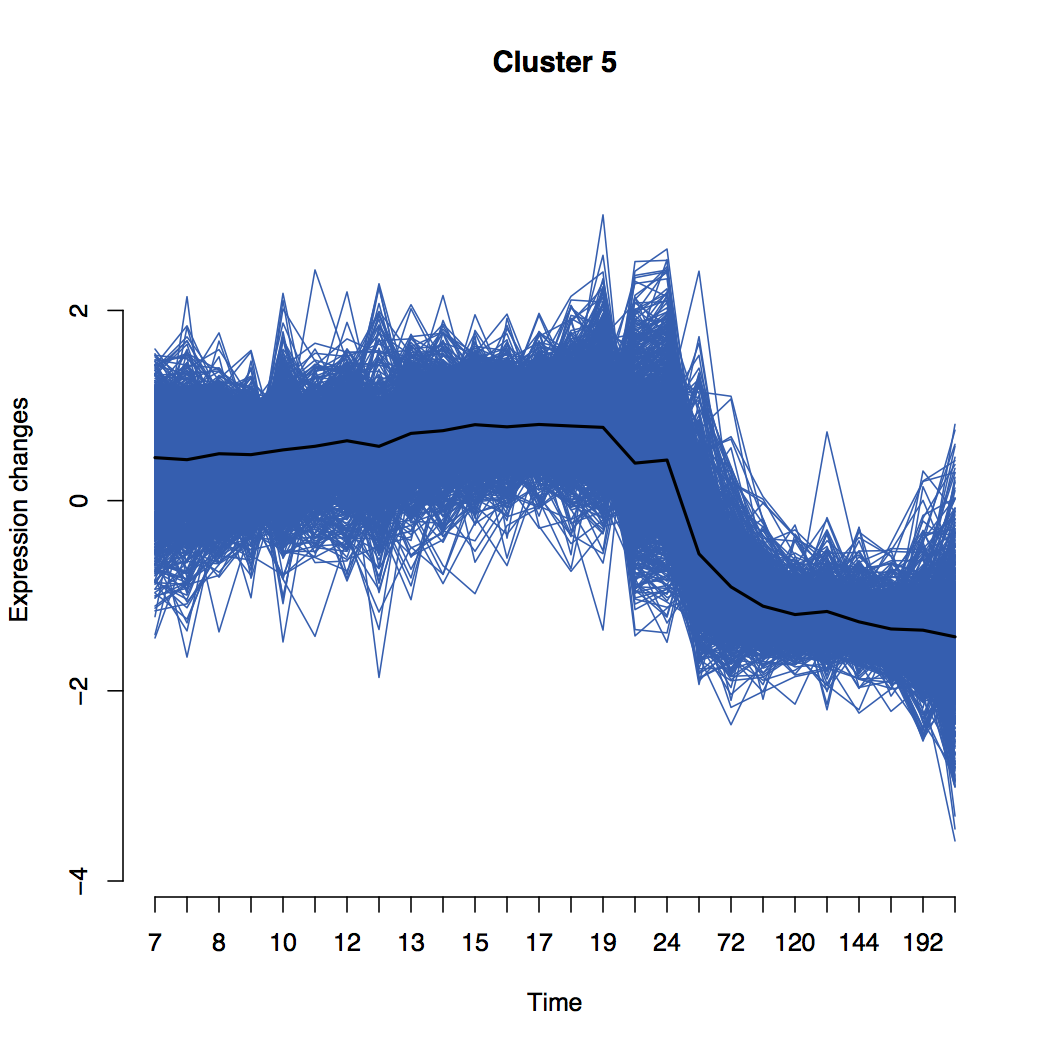
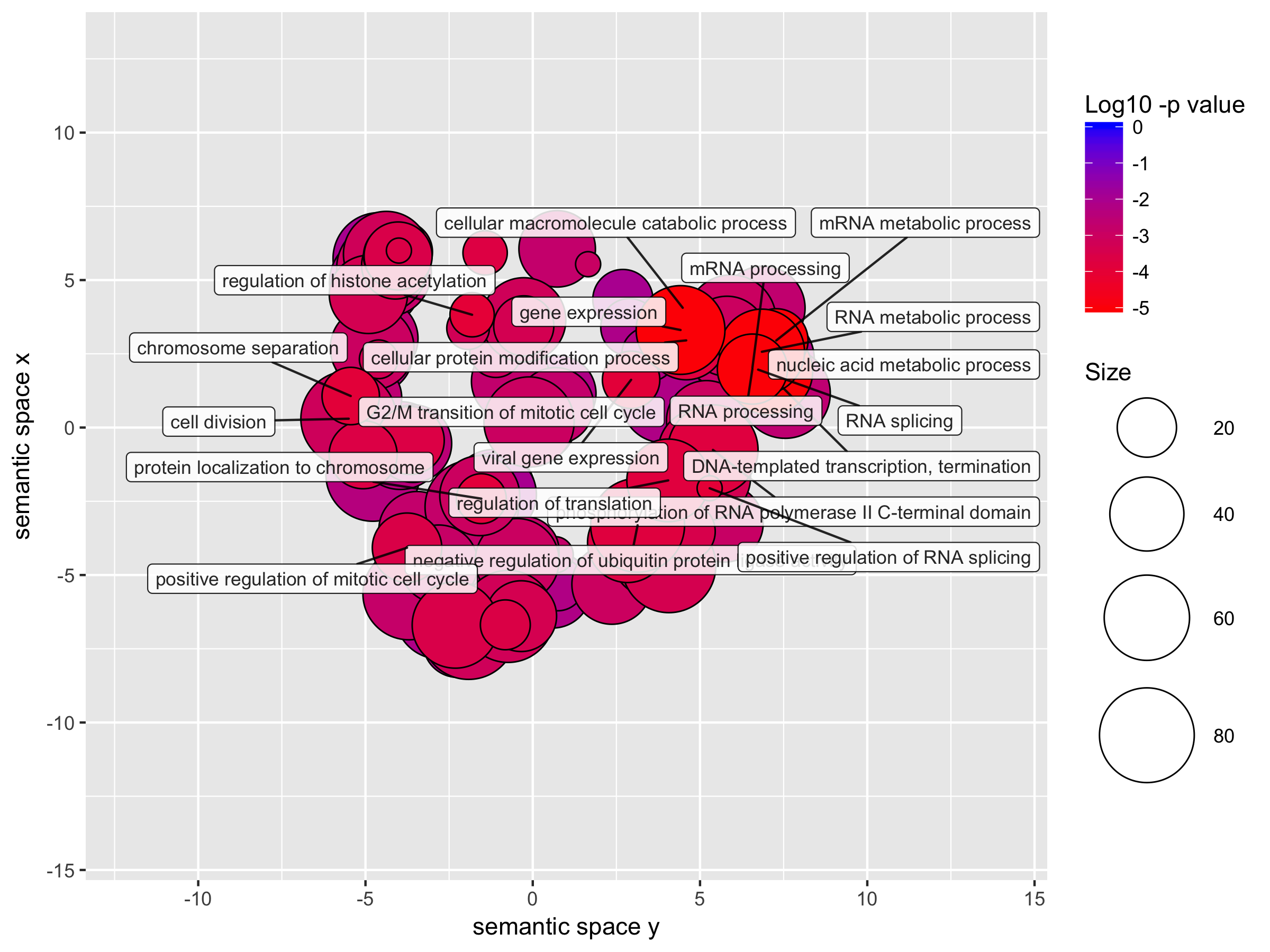
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.166230 | R-6 | 0.997583083256 | E-5 | 0.999999999867 | Q5R4G2 | Platelet-activating factor acetylhydrolase IB subunit beta | gi|156358713|ref|XP_001624660.1|predicted protein [Nematostella vectensis], 1e-109 |
| other nr_hits | gi|999984633|gb|KXJ23774.1|Serine/arginine repetitive matrix protein 1 [Exaiptasia pallida] 4e-68; gi|1005464864|ref|XP_015769451.1|PREDICTED: serine/arginine repetitive matrix protein 1-like isoform X1 [Acropora digitifera] 5e-58; gi|828206445|ref|XP_004208028.2|PREDICTED: serine/arginine repetitive matrix protein 1-like isoform X1 [Hydra vulgaris] 7e-50; gi|828206448|ref|XP_012556964.1|PREDICTED: serine/arginine repetitive matrix protein 1-like isoform X2 [Hydra vulgaris] 7e-50; gi|828193356|ref|XP_012563030.1|PREDICTED: platelet-activating factor acetylhydrolase IB subunit gamma-like [Hydra vulgaris] 4e-45; gi|999986006|gb|KXJ25147.1|Platelet-activating factor acetylhydrolase IB subunit gamma [Exaiptasia pallida] 1e-32; gi|632977719|ref|XP_007905503.1|PREDICTED: platelet-activating factor acetylhydrolase IB subunit beta [Callorhinchus milii] 2e-32; gi|640808203|ref|XP_008060621.1|PREDICTED: platelet-activating factor acetylhydrolase IB subunit gamma [Carlito syrichta] 8e-32; gi|16758470|ref|NP_446106.1|platelet-activating factor acetylhydrolase IB subunit gamma [Rattus norvegicus] 1e-31; | ||||||
| NvERTx.4.10749 | R-7 | 0.729998902652 | E-5 | 0.999999999866 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.10750 | R-7 | 0.729998902652 | E-5 | 0.999999999866 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.37482 | R-7 | 0.729998902652 | E-5 | 0.999999999866 | O35207 | Cyclin-dependent kinase 2-associated protein 1 | gi|156402347|ref|XP_001639552.1|predicted protein [Nematostella vectensis], 8e-27 |
| other nr_hits | gi|970879062|ref|XP_015123452.1|PREDICTED: cyclin-dependent kinase 2-associated protein 2-like [Diachasma alloeum] 8e-19; gi|1049263747|ref|XP_017571815.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like [Pygocentrus nattereri] 1e-18; gi|225706114|gb|ACO08903.1|Cyclin-dependent kinase 2-associated protein 1 [Osmerus mordax] 1e-18; gi|1101531968|ref|XP_018931747.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X1 [Cyprinus carpio] 4e-18; gi|1101531972|ref|XP_018931750.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X3 [Cyprinus carpio] 4e-18; gi|1069780919|ref|XP_018324673.1|PREDICTED: cyclin-dependent kinase 2-associated protein 2 [Agrilus planipennis] 4e-18; gi|1101531964|ref|XP_018931745.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X1 [Cyprinus carpio] 5e-18; gi|1101531966|ref|XP_018931746.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X2 [Cyprinus carpio] 5e-18; gi|1025318772|ref|XP_016386128.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like [Sinocyclocheilus rhinocerous] 5e-18; | ||||||
| NvERTx.4.37483 | R-7 | 0.729998902652 | E-5 | 0.999999999866 | O35207 | Cyclin-dependent kinase 2-associated protein 1 | gi|156402347|ref|XP_001639552.1|predicted protein [Nematostella vectensis], 8e-27 |
| other nr_hits | gi|970879062|ref|XP_015123452.1|PREDICTED: cyclin-dependent kinase 2-associated protein 2-like [Diachasma alloeum] 8e-19; gi|1049263747|ref|XP_017571815.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like [Pygocentrus nattereri] 1e-18; gi|225706114|gb|ACO08903.1|Cyclin-dependent kinase 2-associated protein 1 [Osmerus mordax] 1e-18; gi|1101531968|ref|XP_018931747.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X1 [Cyprinus carpio] 4e-18; gi|1101531972|ref|XP_018931750.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X3 [Cyprinus carpio] 4e-18; gi|1069780919|ref|XP_018324673.1|PREDICTED: cyclin-dependent kinase 2-associated protein 2 [Agrilus planipennis] 4e-18; gi|1101531964|ref|XP_018931745.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X1 [Cyprinus carpio] 5e-18; gi|1101531966|ref|XP_018931746.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like isoform X2 [Cyprinus carpio] 5e-18; gi|1025318772|ref|XP_016386128.1|PREDICTED: cyclin-dependent kinase 2-associated protein 1-like [Sinocyclocheilus rhinocerous] 5e-18; | ||||||
| NvERTx.4.112230 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | Q08BS4 | Histone-lysine N-methyltransferase EZH2 | gi|942313227|gb|JAN38546.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna], 9e-16 |
| other nr_hits | gi|942332913|gb|JAN48389.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942332915|gb|JAN48390.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942347206|gb|JAN55534.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942347208|gb|JAN55535.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942373107|gb|JAN68483.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942341497|gb|JAN52681.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|1022773171|gb|KZS17283.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|1005470515|ref|XP_015772139.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Acropora digitifera] 9e-16; gi|1005482818|ref|XP_015778100.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like, partial [Acropora digitifera] 9e-16; | ||||||
| NvERTx.4.112231 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | Q28D84 | Histone-lysine N-methyltransferase EZH2 | gi|156351239|ref|XP_001622422.1|predicted protein [Nematostella vectensis], 1e-240 |
| other nr_hits | gi|999984762|gb|KXJ23903.1|Histone-lysine N-methyltransferase EZH2 [Exaiptasia pallida] 4e-145; gi|918337546|gb|KOF99173.1|hypothetical protein OCBIM_22018841mg [Octopus bimaculoides] 2e-87; gi|1129951849|ref|XP_019735428.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Hippocampus comes] 9e-87; gi|1149013937|pdb|5LS6|65|||2017|2|22|Chain A, Crystal Structure Of Eed In Complex With A Trimethylated Jarid2 Peptide >pdb|5HYN|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|G Chain G, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|L Chain L, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|R Chain R, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5LS6|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|E Chain E, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|H Chain H, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|K Chain K, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor 6e-86; gi|410927554|ref|XP_003977206.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Takifugu rubripes] 3e-83; gi|1126159730|ref|XP_019621342.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Branchiostoma belcheri] 3e-82; gi|961070973|ref|XP_014777363.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Octopus bimaculoides] 2e-80; gi|1139806135|ref|XP_019923809.1|PREDICTED: histone-lysine N-methyltransferase EZH2 isoform X1 [Crassostrea gigas] 4e-80; gi|1139806137|ref|XP_019923810.1|PREDICTED: histone-lysine N-methyltransferase EZH2 isoform X2 [Crassostrea gigas] 4e-80; | ||||||
| NvERTx.4.112234 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | Q4R381 | Histone-lysine N-methyltransferase EZH2 | gi|156351239|ref|XP_001622422.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999984762|gb|KXJ23903.1|Histone-lysine N-methyltransferase EZH2 [Exaiptasia pallida] 2e-225; gi|918337546|gb|KOF99173.1|hypothetical protein OCBIM_22018841mg [Octopus bimaculoides] 3e-160; gi|1129951849|ref|XP_019735428.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Hippocampus comes] 6e-159; gi|1149013937|pdb|5LS6|65|||2017|2|22|Chain A, Crystal Structure Of Eed In Complex With A Trimethylated Jarid2 Peptide >pdb|5HYN|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|G Chain G, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|L Chain L, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|R Chain R, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5LS6|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|E Chain E, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|H Chain H, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|K Chain K, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor 1e-157; gi|1005470515|ref|XP_015772139.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Acropora digitifera] 8e-156; gi|410927554|ref|XP_003977206.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Takifugu rubripes] 7e-155; gi|1126159730|ref|XP_019621342.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Branchiostoma belcheri] 2e-153; gi|961070973|ref|XP_014777363.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Octopus bimaculoides] 2e-153; gi|524879179|ref|XP_005096267.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like isoform X2 [Aplysia californica] 6e-151; | ||||||
| NvERTx.4.112235 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | Q28D84 | Histone-lysine N-methyltransferase EZH2 | gi|156351239|ref|XP_001622422.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999984762|gb|KXJ23903.1|Histone-lysine N-methyltransferase EZH2 [Exaiptasia pallida] 3e-286; gi|1005470515|ref|XP_015772139.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Acropora digitifera] 3e-217; gi|1129951849|ref|XP_019735428.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Hippocampus comes] 6e-214; gi|1149013937|pdb|5LS6|65|||2017|2|22|Chain A, Crystal Structure Of Eed In Complex With A Trimethylated Jarid2 Peptide >pdb|5HYN|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|G Chain G, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|L Chain L, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|R Chain R, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5LS6|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|E Chain E, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|H Chain H, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|K Chain K, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor 8e-213; gi|410927554|ref|XP_003977206.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Takifugu rubripes] 4e-210; gi|1126159730|ref|XP_019621342.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Branchiostoma belcheri] 3e-209; gi|524879179|ref|XP_005096267.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like isoform X2 [Aplysia californica] 2e-205; gi|1139806135|ref|XP_019923809.1|PREDICTED: histone-lysine N-methyltransferase EZH2 isoform X1 [Crassostrea gigas] 2e-205; gi|1139806137|ref|XP_019923810.1|PREDICTED: histone-lysine N-methyltransferase EZH2 isoform X2 [Crassostrea gigas] 2e-205; | ||||||
| NvERTx.4.112236 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | Q08BS4 | Histone-lysine N-methyltransferase EZH2 | gi|942313227|gb|JAN38546.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna], 9e-16 |
| other nr_hits | gi|942332913|gb|JAN48389.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942332915|gb|JAN48390.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942347206|gb|JAN55534.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942347208|gb|JAN55535.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942373107|gb|JAN68483.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|942341497|gb|JAN52681.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|1022773171|gb|KZS17283.1|Histone-lysine N-methyltransferase E(z) [Daphnia magna] 9e-16; gi|1005470515|ref|XP_015772139.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Acropora digitifera] 9e-16; gi|1005482818|ref|XP_015778100.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like, partial [Acropora digitifera] 9e-16; | ||||||
| NvERTx.4.112237 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | Q4R381 | Histone-lysine N-methyltransferase EZH2 | gi|156351239|ref|XP_001622422.1|predicted protein [Nematostella vectensis], 5e-270 |
| other nr_hits | gi|999984762|gb|KXJ23903.1|Histone-lysine N-methyltransferase EZH2 [Exaiptasia pallida] 4e-174; gi|1129951849|ref|XP_019735428.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Hippocampus comes] 1e-110; gi|918337546|gb|KOF99173.1|hypothetical protein OCBIM_22018841mg [Octopus bimaculoides] 4e-110; gi|1149013937|pdb|5LS6|65|||2017|2|22|Chain A, Crystal Structure Of Eed In Complex With A Trimethylated Jarid2 Peptide >pdb|5HYN|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|G Chain G, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|L Chain L, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5HYN|R Chain R, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Oncogenic Histone H3k27m Peptide >pdb|5LS6|B Chain B, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|E Chain E, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|H Chain H, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor >pdb|5LS6|K Chain K, Structure Of Human Polycomb Repressive Complex 2 (prc2) With Inhibitor 2e-109; gi|410927554|ref|XP_003977206.1|PREDICTED: histone-lysine N-methyltransferase EZH2 [Takifugu rubripes] 5e-107; gi|1126159730|ref|XP_019621342.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Branchiostoma belcheri] 1e-106; gi|1005470515|ref|XP_015772139.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Acropora digitifera] 6e-106; gi|1048454504|ref|XP_017499233.1|PREDICTED: histone-lysine N-methyltransferase EZH1 isoform X3 [Manis javanica] 9e-104; gi|926623582|ref|XP_013777216.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Limulus polyphemus] 2e-103; | ||||||
| NvERTx.4.112228 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.112232 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.112233 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.213333 | R-7 | 0.942298898567 | E-5 | 0.999999999865 | P70351 | Histone-lysine N-methyltransferase EZH1 | gi|156351239|ref|XP_001622422.1|predicted protein [Nematostella vectensis], 3e-21 |
| other nr_hits | gi|999984762|gb|KXJ23903.1|Histone-lysine N-methyltransferase EZH2 [Exaiptasia pallida] 6e-07; gi|1005470515|ref|XP_015772139.1|PREDICTED: histone-lysine N-methyltransferase EZH2-like [Acropora digitifera] 8e-06; | ||||||
| NvERTx.4.232116 | R-5 | 0.720165835684 | E-5 | 0.999999999861 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.94505 | R-5 | 0.720165835684 | E-5 | 0.999999999861 | Q5T5N4 | Uncharacterized protein C6orf118 | gi|1005445430|ref|XP_015760014.1|PREDICTED: uncharacterized protein C6orf118-like [Acropora digitifera], 2e-115 |
| other nr_hits | gi|999972215|gb|KXJ11389.1|Uncharacterized protein C6orf118 [Exaiptasia pallida] 2e-91; gi|1126211098|ref|XP_019639327.1|PREDICTED: uncharacterized protein C6orf118-like [Branchiostoma belcheri] 4e-62; gi|291225834|ref|XP_002732903.1|PREDICTED: uncharacterized protein C6orf118-like [Saccoglossus kowalevskii] 4e-62; gi|260790707|ref|XP_002590383.1|hypothetical protein BRAFLDRAFT_76659 [Branchiostoma floridae] 9e-61; gi|919064746|ref|XP_013414010.1|PREDICTED: uncharacterized protein LOC106176247 [Lingula anatina] 4e-56; gi|762130744|ref|XP_011448514.1|PREDICTED: uncharacterized protein C6orf118 [Crassostrea gigas] 5e-50; gi|390345733|ref|XP_003726397.1|PREDICTED: uncharacterized protein C6orf118 [Strongylocentrotus purpuratus] 9e-50; gi|524866518|ref|XP_005090080.1|PREDICTED: uncharacterized protein C6orf118-like isoform X2 [Aplysia californica] 3e-44; gi|443734125|gb|ELU18221.1|hypothetical protein CAPTEDRAFT_221744 [Capitella teleta] 3e-44; | ||||||
| NvERTx.4.194145 | R-5 | 0.720165835684 | E-5 | 0.999999999861 | Q9LJG8 | Trihelix transcription factor ASIL2 {ECO:0000305} | gi|999972211|gb|KXJ11385.1|hypothetical protein AC249_AIPGENE8039 [Exaiptasia pallida], 8e-63 |
| other nr_hits | gi|1005445395|ref|XP_015759999.1|PREDICTED: uncharacterized protein LOC107339249 [Acropora digitifera] 8e-34; gi|156375294|ref|XP_001630016.1|predicted protein [Nematostella vectensis] 3e-11; gi|1005432527|ref|XP_015753830.1|PREDICTED: uncharacterized protein LOC107333516 [Acropora digitifera] 4e-10; gi|999981599|gb|KXJ20773.1|hypothetical protein AC249_AIPGENE421 [Exaiptasia pallida] 3e-09; gi|999981466|gb|KXJ20640.1|hypothetical protein AC249_AIPGENE941 [Exaiptasia pallida] 5e-09; gi|999967536|gb|KXJ06710.1|Trihelix transcription factor GT-3a [Exaiptasia pallida] 1e-07; gi|999970446|gb|KXJ09620.1|hypothetical protein AC249_AIPGENE23032 [Exaiptasia pallida] 7e-06; gi|672138796|ref|XP_008793163.1|PREDICTED: trihelix transcription factor ASIL2-like [Phoenix dactylifera] 3e-05; | ||||||
| NvERTx.4.194146 | R-5 | 0.720165835684 | E-5 | 0.999999999861 | Q9LJG8 | Trihelix transcription factor ASIL2 {ECO:0000305} | gi|999972211|gb|KXJ11385.1|hypothetical protein AC249_AIPGENE8039 [Exaiptasia pallida], 1e-62 |
| other nr_hits | gi|1005445395|ref|XP_015759999.1|PREDICTED: uncharacterized protein LOC107339249 [Acropora digitifera] 8e-34; gi|156375294|ref|XP_001630016.1|predicted protein [Nematostella vectensis] 3e-11; gi|1005432527|ref|XP_015753830.1|PREDICTED: uncharacterized protein LOC107333516 [Acropora digitifera] 4e-10; gi|999981599|gb|KXJ20773.1|hypothetical protein AC249_AIPGENE421 [Exaiptasia pallida] 3e-09; gi|999981466|gb|KXJ20640.1|hypothetical protein AC249_AIPGENE941 [Exaiptasia pallida] 5e-09; gi|999967536|gb|KXJ06710.1|Trihelix transcription factor GT-3a [Exaiptasia pallida] 1e-07; gi|999970446|gb|KXJ09620.1|hypothetical protein AC249_AIPGENE23032 [Exaiptasia pallida] 7e-06; gi|672138796|ref|XP_008793163.1|PREDICTED: trihelix transcription factor ASIL2-like [Phoenix dactylifera] 3e-05; | ||||||
| NvERTx.4.194147 | R-5 | 0.720165835684 | E-5 | 0.999999999861 | Q5R9W6 | Transcription initiation factor TFIID subunit 13 | gi|156384029|ref|XP_001633134.1|predicted protein [Nematostella vectensis], 7e-43 |
| other nr_hits | gi|999972223|gb|KXJ11397.1|Transcription initiation factor TFIID subunit 13 [Exaiptasia pallida] 4e-41; gi|221120854|ref|XP_002157895.1|PREDICTED: transcription initiation factor TFIID subunit 13-like [Hydra vulgaris] 3e-34; gi|951532437|ref|XP_014470088.1|PREDICTED: transcription initiation factor TFIID subunit 13 isoform X2 [Dinoponera quadriceps] 3e-32; gi|766922439|ref|XP_011505792.1|PREDICTED: transcription initiation factor TFIID subunit 13 [Ceratosolen solmsi marchali] 3e-32; gi|665797989|ref|XP_008546849.1|PREDICTED: transcription initiation factor TFIID subunit 13 isoform X2 [Microplitis demolitor] 3e-32; gi|1025002914|ref|XP_016320450.1|PREDICTED: transcription initiation factor TFIID subunit 13 [Sinocyclocheilus anshuiensis] 7e-32; gi|1020467232|ref|XP_016138602.1|PREDICTED: transcription initiation factor TFIID subunit 13 [Sinocyclocheilus grahami] 7e-32; gi|966653025|gb|KTF87927.1|hypothetical protein cypCar_00005595 [Cyprinus carpio] 7e-32; gi|1061115522|ref|XP_017883960.1|PREDICTED: transcription initiation factor TFIID subunit 13 isoform X2 [Ceratina calcarata] 9e-32; | ||||||
| NvERTx.4.194148 | R-5 | 0.720165835684 | E-5 | 0.999999999861 | Q9LJG8 | Trihelix transcription factor ASIL2 {ECO:0000305} | gi|999972211|gb|KXJ11385.1|hypothetical protein AC249_AIPGENE8039 [Exaiptasia pallida], 1e-62 |
| other nr_hits | gi|1005445395|ref|XP_015759999.1|PREDICTED: uncharacterized protein LOC107339249 [Acropora digitifera] 8e-34; gi|156375294|ref|XP_001630016.1|predicted protein [Nematostella vectensis] 3e-11; gi|1005432527|ref|XP_015753830.1|PREDICTED: uncharacterized protein LOC107333516 [Acropora digitifera] 4e-10; gi|999981599|gb|KXJ20773.1|hypothetical protein AC249_AIPGENE421 [Exaiptasia pallida] 3e-09; gi|999981466|gb|KXJ20640.1|hypothetical protein AC249_AIPGENE941 [Exaiptasia pallida] 5e-09; gi|999967536|gb|KXJ06710.1|Trihelix transcription factor GT-3a [Exaiptasia pallida] 1e-07; gi|999970446|gb|KXJ09620.1|hypothetical protein AC249_AIPGENE23032 [Exaiptasia pallida] 7e-06; gi|672138796|ref|XP_008793163.1|PREDICTED: trihelix transcription factor ASIL2-like [Phoenix dactylifera] 3e-05; | ||||||
| NvERTx.4.194149 | R-5 | 0.720165835684 | E-5 | 0.999999999861 | Q5R9W6 | Transcription initiation factor TFIID subunit 13 | gi|156384029|ref|XP_001633134.1|predicted protein [Nematostella vectensis], 1e-52 |
| other nr_hits | gi|999972223|gb|KXJ11397.1|Transcription initiation factor TFIID subunit 13 [Exaiptasia pallida] 2e-49; gi|221120854|ref|XP_002157895.1|PREDICTED: transcription initiation factor TFIID subunit 13-like [Hydra vulgaris] 8e-42; gi|1025002914|ref|XP_016320450.1|PREDICTED: transcription initiation factor TFIID subunit 13 [Sinocyclocheilus anshuiensis] 6e-39; gi|1020467232|ref|XP_016138602.1|PREDICTED: transcription initiation factor TFIID subunit 13 [Sinocyclocheilus grahami] 6e-39; gi|966653025|gb|KTF87927.1|hypothetical protein cypCar_00005595 [Cyprinus carpio] 6e-39; gi|1048466934|ref|XP_017513447.1|PREDICTED: transcription initiation factor TFIID subunit 13 [Manis javanica] 1e-38; gi|665797989|ref|XP_008546849.1|PREDICTED: transcription initiation factor TFIID subunit 13 isoform X2 [Microplitis demolitor] 1e-38; gi|557014625|ref|XP_006007810.1|PREDICTED: transcription initiation factor TFIID subunit 13 [Latimeria chalumnae] 1e-38; gi|899135817|ref|NP_001297979.1|transcription initiation factor TFIID subunit 13 [Esox lucius] 1e-38; | ||||||
| NvERTx.4.31866 | R-5 | 0.999985803381 | E-5 | 0.999999999861 | No_Uniprotmatch | No_Description | gi|156371461|ref|XP_001628782.1|predicted protein [Nematostella vectensis], 2e-32 |
| other nr_hits | NA | ||||||
| NvERTx.4.43452 | R-5 | 0.999985803381 | E-5 | 0.999999999861 | K1Q279_CRAGI | Targeting protein for Xklp2 {ECO:0000313|EMBL:EKC25469.1} | gi|156371461|ref|XP_001628782.1|predicted protein [Nematostella vectensis], 7e-154 |
| other nr_hits | gi|762073488|ref|XP_011433915.1|PREDICTED: targeting protein for Xklp2 homolog isoform X1 [Crassostrea gigas] 1e-05; gi|762073490|ref|XP_011433925.1|PREDICTED: targeting protein for Xklp2 isoform X2 [Crassostrea gigas] 1e-05; gi|405959427|gb|EKC25469.1|Targeting protein for Xklp2 [Crassostrea gigas] 1e-05; gi|1005437882|ref|XP_015756372.1|PREDICTED: targeting protein for Xklp2 homolog [Acropora digitifera] 2e-05; gi|408407565|sp|TPX2_PATPE|E2RYF8.1|RecName: Full=Targeting protein for Xklp2 homolog; Short=TPX2 homolog; AltName: Full=Microtubule-associated protein TPX2 homolog >BAJ24842.1 microtubule-associated protein homolog-like [Patiria pectinifera] 2e-05; | ||||||
| NvERTx.4.43453 | R-5 | 0.999985803381 | E-5 | 0.999999999861 | E2RYF8 | Targeting protein for Xklp2 homolog | gi|156371461|ref|XP_001628782.1|predicted protein [Nematostella vectensis], 4e-294 |
| other nr_hits | gi|999988272|gb|KXJ27366.1|Targeting protein for Xklp2-like [Exaiptasia pallida] 1e-129; gi|1005437882|ref|XP_015756372.1|PREDICTED: targeting protein for Xklp2 homolog [Acropora digitifera] 2e-106; gi|156371459|ref|XP_001628781.1|predicted protein [Nematostella vectensis] 8e-93; gi|999988280|gb|KXJ27374.1|Targeting protein for Xklp2 [Exaiptasia pallida] 2e-90; gi|919017960|ref|XP_013393247.1|PREDICTED: targeting protein for Xklp2 homolog [Lingula anatina] 3e-75; gi|1126209226|ref|XP_019638315.1|PREDICTED: LOW QUALITY PROTEIN: targeting protein for Xklp2 homolog [Branchiostoma belcheri] 7e-75; gi|260798154|ref|XP_002594065.1|hypothetical protein BRAFLDRAFT_68495 [Branchiostoma floridae] 3e-74; gi|408407565|sp|TPX2_PATPE|E2RYF8.1|RecName: Full=Targeting protein for Xklp2 homolog; Short=TPX2 homolog; AltName: Full=Microtubule-associated protein TPX2 homolog >BAJ24842.1 microtubule-associated protein homolog-like [Patiria pectinifera] 8e-74; gi|449671026|ref|XP_002162579.2|PREDICTED: targeting protein for Xklp2 homolog [Hydra vulgaris] 3e-72; | ||||||
| NvERTx.4.109319 | R-8 | 0.72466836489 | E-5 | 0.999999999858 | O15063 | Uncharacterized protein KIAA0355 | gi|1005459631|ref|XP_015766890.1|PREDICTED: uncharacterized protein KIAA0355-like [Acropora digitifera], 8e-176 |
| other nr_hits | gi|999982974|gb|KXJ22120.1|Uncharacterized protein KIAA0355 [Exaiptasia pallida] 3e-175; gi|195998291|ref|XP_002109014.1|predicted protein [Trichoplax adhaerens] 5e-46; gi|340374429|ref|XP_003385740.1|PREDICTED: uncharacterized protein LOC100640706 [Amphimedon queenslandica] 2e-32; gi|395505801|ref|XP_003757226.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Sarcophilus harrisii] 3e-22; gi|126296003|ref|XP_001365227.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Monodelphis domestica] 5e-22; gi|975102767|ref|XP_015266588.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Gekko japonicus] 6e-22; gi|768934873|ref|XP_011607872.1|PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein KIAA0355 homolog [Takifugu rubripes] 1e-21; gi|1117271222|ref|XP_019361110.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Gavialis gangeticus] 3e-21; gi|1025360667|ref|XP_016402296.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus rhinocerous] 3e-21; | ||||||
| NvERTx.4.109320 | R-8 | 0.72466836489 | E-5 | 0.999999999858 | O15063 | Uncharacterized protein KIAA0355 | gi|999982974|gb|KXJ22120.1|Uncharacterized protein KIAA0355 [Exaiptasia pallida], 2e-197 |
| other nr_hits | gi|1005459631|ref|XP_015766890.1|PREDICTED: uncharacterized protein KIAA0355-like [Acropora digitifera] 3e-91; gi|449665043|ref|XP_002166712.2|PREDICTED: uncharacterized protein LOC100202097 [Hydra vulgaris] 2e-22; gi|195998291|ref|XP_002109014.1|predicted protein [Trichoplax adhaerens] 1e-16; gi|1020469671|ref|XP_016139902.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus grahami] 1e-16; gi|966652471|gb|KTF87373.1|hypothetical protein cypCar_00024528, partial [Cyprinus carpio] 1e-16; gi|1025360667|ref|XP_016402296.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus rhinocerous] 2e-16; gi|1025266512|ref|XP_016373381.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus rhinocerous] 2e-16; gi|1101556677|ref|XP_018944292.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Cyprinus carpio] 3e-16; gi|1024991389|ref|XP_016315826.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus anshuiensis] 3e-16; | ||||||
| NvERTx.4.109322 | R-8 | 0.72466836489 | E-5 | 0.999999999858 | O15063 | Uncharacterized protein KIAA0355 | gi|999982974|gb|KXJ22120.1|Uncharacterized protein KIAA0355 [Exaiptasia pallida], 0e+00 |
| other nr_hits | gi|1005459631|ref|XP_015766890.1|PREDICTED: uncharacterized protein KIAA0355-like [Acropora digitifera] 3e-231; gi|195998291|ref|XP_002109014.1|predicted protein [Trichoplax adhaerens] 8e-61; gi|340374429|ref|XP_003385740.1|PREDICTED: uncharacterized protein LOC100640706 [Amphimedon queenslandica] 8e-36; gi|1101556677|ref|XP_018944292.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Cyprinus carpio] 9e-35; gi|1025360667|ref|XP_016402296.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus rhinocerous] 1e-34; gi|432853115|ref|XP_004067547.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Oryzias latipes] 1e-34; gi|1025266512|ref|XP_016373381.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus rhinocerous] 2e-34; gi|1020469671|ref|XP_016139902.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus grahami] 3e-34; gi|966709571|gb|KTG41809.1|hypothetical protein cypCar_00006843 [Cyprinus carpio] 5e-34; | ||||||
| NvERTx.4.109325 | R-8 | 0.72466836489 | E-5 | 0.999999999858 | O15063 | Uncharacterized protein KIAA0355 | gi|999982974|gb|KXJ22120.1|Uncharacterized protein KIAA0355 [Exaiptasia pallida], 2e-197 |
| other nr_hits | gi|1005459631|ref|XP_015766890.1|PREDICTED: uncharacterized protein KIAA0355-like [Acropora digitifera] 3e-91; gi|449665043|ref|XP_002166712.2|PREDICTED: uncharacterized protein LOC100202097 [Hydra vulgaris] 2e-22; gi|195998291|ref|XP_002109014.1|predicted protein [Trichoplax adhaerens] 1e-16; gi|1020469671|ref|XP_016139902.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus grahami] 1e-16; gi|966652471|gb|KTF87373.1|hypothetical protein cypCar_00024528, partial [Cyprinus carpio] 1e-16; gi|1025360667|ref|XP_016402296.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus rhinocerous] 2e-16; gi|1025266512|ref|XP_016373381.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus rhinocerous] 2e-16; gi|1101556677|ref|XP_018944292.1|PREDICTED: uncharacterized protein KIAA0355 homolog [Cyprinus carpio] 3e-16; gi|1024991389|ref|XP_016315826.1|PREDICTED: uncharacterized protein KIAA0355-like [Sinocyclocheilus anshuiensis] 3e-16; | ||||||
| NvERTx.4.3180 | R-5 | 0.575841379563 | E-5 | 0.999999999855 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.134759 | R-5 | 0.575841379563 | E-5 | 0.999999999855 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.134760 | R-5 | 0.575841379563 | E-5 | 0.999999999855 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.134762 | R-5 | 0.575841379563 | E-5 | 0.999999999855 | Q12996 | Cleavage stimulation factor subunit 3 | gi|156383316|ref|XP_001632780.1|predicted protein [Nematostella vectensis], 7e-46 |
| other nr_hits | gi|1005439757|ref|XP_015757236.1|PREDICTED: cleavage stimulation factor subunit 3-like isoform X1 [Acropora digitifera] 6e-27; gi|1005439760|ref|XP_015757237.1|PREDICTED: cleavage stimulation factor subunit 3-like isoform X2 [Acropora digitifera] 6e-27; gi|999986898|gb|KXJ26025.1|Cleavage stimulation factor subunit 3 [Exaiptasia pallida] 7e-25; gi|828216948|ref|XP_012560497.1|PREDICTED: cleavage stimulation factor subunit 3-like [Hydra vulgaris] 1e-16; gi|196001179|ref|XP_002110457.1|hypothetical protein TRIADDRAFT_21977 [Trichoplax adhaerens] 3e-15; gi|1133450135|ref|XP_019857540.1|PREDICTED: cleavage stimulation factor subunit 3 [Amphimedon queenslandica] 4e-14; gi|1062801519|ref|XP_017945601.1|PREDICTED: cleavage stimulation factor subunit 3-like [Xenopus tropicalis] 6e-14; gi|1062801516|ref|XP_017945600.1|PREDICTED: cleavage stimulation factor subunit 3-like [Xenopus tropicalis] 1e-13; gi|1062881973|ref|XP_017948669.1|PREDICTED: cleavage stimulation factor subunit 3-like [Xenopus tropicalis] 2e-13; | ||||||
| NvERTx.4.134763 | R-5 | 0.575841379563 | E-5 | 0.999999999855 | Q5RDW9 | Cleavage stimulation factor subunit 3 | gi|156383316|ref|XP_001632780.1|predicted protein [Nematostella vectensis], 5e-103 |
| other nr_hits | gi|999984586|gb|KXJ23727.1|Cleavage stimulation factor subunit 3 [Exaiptasia pallida] 8e-75; gi|1005439757|ref|XP_015757236.1|PREDICTED: cleavage stimulation factor subunit 3-like isoform X1 [Acropora digitifera] 2e-60; gi|1005439760|ref|XP_015757237.1|PREDICTED: cleavage stimulation factor subunit 3-like isoform X2 [Acropora digitifera] 4e-59; gi|585707460|ref|XP_006823989.1|PREDICTED: LOW QUALITY PROTEIN: cleavage stimulation factor subunit 3-like [Saccoglossus kowalevskii] 8e-46; gi|926644767|ref|XP_013788661.1|PREDICTED: LOW QUALITY PROTEIN: cleavage stimulation factor subunit 3-like [Limulus polyphemus] 1e-44; gi|902887105|ref|XP_013033143.1|PREDICTED: cleavage stimulation factor subunit 3 [Anser cygnoides domesticus] 1e-40; gi|874480661|ref|XP_012957401.1|PREDICTED: cleavage stimulation factor subunit 3 [Anas platyrhynchos] 1e-40; gi|483510128|gb|EOA99519.1|Cleavage stimulation factor 77 kDa subunit, partial [Anas platyrhynchos] 1e-40; gi|632941608|ref|XP_007885954.1|PREDICTED: cleavage stimulation factor subunit 3 isoform X1 [Callorhinchus milii] 2e-40; | ||||||
| NvERTx.4.134764 | R-5 | 0.575841379563 | E-5 | 0.999999999855 | Q12996 | Cleavage stimulation factor subunit 3 | gi|156383316|ref|XP_001632780.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|1005439757|ref|XP_015757236.1|PREDICTED: cleavage stimulation factor subunit 3-like isoform X1 [Acropora digitifera] 5e-285; gi|1005439760|ref|XP_015757237.1|PREDICTED: cleavage stimulation factor subunit 3-like isoform X2 [Acropora digitifera] 9e-284; gi|999984586|gb|KXJ23727.1|Cleavage stimulation factor subunit 3 [Exaiptasia pallida] 9e-279; gi|585707460|ref|XP_006823989.1|PREDICTED: LOW QUALITY PROTEIN: cleavage stimulation factor subunit 3-like [Saccoglossus kowalevskii] 1e-234; gi|632941610|ref|XP_007885955.1|PREDICTED: cleavage stimulation factor subunit 3 isoform X2 [Callorhinchus milii] 5e-226; gi|704532077|ref|XP_010181609.1|PREDICTED: cleavage stimulation factor subunit 3 [Mesitornis unicolor] 2e-225; gi|677424510|gb|KFQ27149.1|Cleavage stimulation factor subunit 3, partial [Mesitornis unicolor] 2e-225; gi|902887105|ref|XP_013033143.1|PREDICTED: cleavage stimulation factor subunit 3 [Anser cygnoides domesticus] 3e-225; gi|874480661|ref|XP_012957401.1|PREDICTED: cleavage stimulation factor subunit 3 [Anas platyrhynchos] 3e-225; | ||||||
| NvERTx.4.172003 | R-5 | 0.575841379563 | E-5 | 0.999999999855 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.14799 | R-8 | 0.999431628617 | E-5 | 0.999999999853 | Q5PPV3 | MORN repeat-containing protein 3 | gi|156384228|ref|XP_001633233.1|predicted protein [Nematostella vectensis], 3e-16 |
| other nr_hits | gi|999976803|gb|KXJ15977.1|MORN repeat-containing protein 3 [Exaiptasia pallida] 2e-13; gi|1005430025|ref|XP_015752760.1|PREDICTED: MORN repeat-containing protein 3-like [Acropora digitifera] 2e-13; gi|443709463|gb|ELU04135.1|hypothetical protein CAPTEDRAFT_167723 [Capitella teleta] 5e-12; gi|918998145|ref|XP_013395207.1|PREDICTED: MORN repeat-containing protein 3-like [Lingula anatina] 8e-11; gi|919000225|ref|XP_013403272.1|PREDICTED: MORN repeat-containing protein 3-like [Lingula anatina] 8e-11; gi|762091087|ref|XP_011427931.1|PREDICTED: MORN repeat-containing protein 3 [Crassostrea gigas] 8e-11; gi|195996933|ref|XP_002108335.1|hypothetical protein TRIADDRAFT_19040 [Trichoplax adhaerens] 8e-11; gi|405976919|gb|EKC41397.1|MORN repeat-containing protein 3 [Crassostrea gigas] 8e-11; gi|260803459|ref|XP_002596607.1|hypothetical protein BRAFLDRAFT_280249 [Branchiostoma floridae] 2e-10; | ||||||
| NvERTx.4.14800 | R-8 | 0.999431628617 | E-5 | 0.999999999853 | Q5PPV3 | MORN repeat-containing protein 3 | gi|405976919|gb|EKC41397.1|MORN repeat-containing protein 3 [Crassostrea gigas], 2e-34 |
| other nr_hits | gi|156384228|ref|XP_001633233.1|predicted protein [Nematostella vectensis] 6e-31; gi|961090401|ref|XP_014771792.1|PREDICTED: MORN repeat-containing protein 3-like [Octopus bimaculoides] 3e-30; gi|1012188282|ref|XP_016009321.1|PREDICTED: MORN repeat-containing protein 3 isoform X3 [Rousettus aegyptiacus] 4e-29; gi|984131808|ref|XP_015357867.1|PREDICTED: MORN repeat-containing protein 3 [Marmota marmota marmota] 6e-29; gi|291406921|ref|XP_002719780.1|PREDICTED: MORN repeat-containing protein 3 [Oryctolagus cuniculus] 1e-28; gi|1003964114|ref|XP_015733623.1|PREDICTED: MORN repeat-containing protein 3 [Coturnix japonica] 2e-28; gi|731266641|ref|XP_010602700.1|PREDICTED: MORN repeat-containing protein 3 isoform X2 [Fukomys damarensis] 3e-28; gi|706127305|ref|XP_010198507.1|PREDICTED: MORN repeat-containing protein 3 [Colius striatus] 3e-28; gi|677064167|gb|KFP25375.1|MORN repeat-containing protein 3, partial [Colius striatus] 3e-28; | ||||||
| NvERTx.4.70854 | R-8 | 0.999431628617 | E-5 | 0.999999999853 | Q5PPV3 | MORN repeat-containing protein 3 | gi|156384228|ref|XP_001633233.1|predicted protein [Nematostella vectensis], 9e-126 |
| other nr_hits | gi|1126158768|ref|XP_019616454.1|PREDICTED: MORN repeat-containing protein 3-like [Branchiostoma belcheri] 2e-95; gi|443709463|gb|ELU04135.1|hypothetical protein CAPTEDRAFT_167723 [Capitella teleta] 1e-90; gi|291233775|ref|XP_002736816.1|PREDICTED: MORN repeat-containing protein 3-like [Saccoglossus kowalevskii] 2e-90; gi|918998145|ref|XP_013395207.1|PREDICTED: MORN repeat-containing protein 3-like [Lingula anatina] 6e-88; gi|762091087|ref|XP_011427931.1|PREDICTED: MORN repeat-containing protein 3 [Crassostrea gigas] 1e-87; gi|942124052|ref|XP_014339910.1|PREDICTED: MORN repeat-containing protein 3 isoform X1 [Latimeria chalumnae] 2e-87; gi|524890245|ref|XP_005101670.1|PREDICTED: MORN repeat-containing protein 3-like [Aplysia californica] 1e-86; gi|405976919|gb|EKC41397.1|MORN repeat-containing protein 3 [Crassostrea gigas] 3e-85; gi|676427966|ref|XP_009045120.1|hypothetical protein LOTGIDRAFT_204987 [Lottia gigantea] 5e-84; | ||||||
| NvERTx.4.70855 | R-8 | 0.999431628617 | E-5 | 0.999999999853 | Q5PPV3 | MORN repeat-containing protein 3 | gi|156384228|ref|XP_001633233.1|predicted protein [Nematostella vectensis], 5e-139 |
| other nr_hits | gi|1126158768|ref|XP_019616454.1|PREDICTED: MORN repeat-containing protein 3-like [Branchiostoma belcheri] 2e-100; gi|291233775|ref|XP_002736816.1|PREDICTED: MORN repeat-containing protein 3-like [Saccoglossus kowalevskii] 2e-97; gi|918998145|ref|XP_013395207.1|PREDICTED: MORN repeat-containing protein 3-like [Lingula anatina] 6e-92; gi|443709463|gb|ELU04135.1|hypothetical protein CAPTEDRAFT_167723 [Capitella teleta] 1e-91; gi|524890245|ref|XP_005101670.1|PREDICTED: MORN repeat-containing protein 3-like [Aplysia californica] 2e-91; gi|942124052|ref|XP_014339910.1|PREDICTED: MORN repeat-containing protein 3 isoform X1 [Latimeria chalumnae] 4e-91; gi|762091087|ref|XP_011427931.1|PREDICTED: MORN repeat-containing protein 3 [Crassostrea gigas] 9e-91; gi|72007501|ref|XP_782568.1|PREDICTED: MORN repeat-containing protein 3 [Strongylocentrotus purpuratus] 3e-89; gi|676427966|ref|XP_009045120.1|hypothetical protein LOTGIDRAFT_204987 [Lottia gigantea] 2e-88; | ||||||
| NvERTx.4.70856 | R-8 | 0.999431628617 | E-5 | 0.999999999853 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.70857 | R-8 | 0.999431628617 | E-5 | 0.999999999853 | Q5PPV3 | MORN repeat-containing protein 3 | gi|156384228|ref|XP_001633233.1|predicted protein [Nematostella vectensis], 5e-85 |
| other nr_hits | gi|1005430025|ref|XP_015752760.1|PREDICTED: MORN repeat-containing protein 3-like [Acropora digitifera] 1e-72; gi|999976803|gb|KXJ15977.1|MORN repeat-containing protein 3 [Exaiptasia pallida] 1e-61; gi|919000225|ref|XP_013403272.1|PREDICTED: MORN repeat-containing protein 3-like [Lingula anatina] 6e-60; gi|524890245|ref|XP_005101670.1|PREDICTED: MORN repeat-containing protein 3-like [Aplysia californica] 5e-59; gi|942124058|ref|XP_014339918.1|PREDICTED: MORN repeat-containing protein 3 isoform X2 [Latimeria chalumnae] 4e-58; gi|676427966|ref|XP_009045120.1|hypothetical protein LOTGIDRAFT_204987 [Lottia gigantea] 4e-57; gi|762091087|ref|XP_011427931.1|PREDICTED: MORN repeat-containing protein 3 [Crassostrea gigas] 2e-56; gi|918998145|ref|XP_013395207.1|PREDICTED: MORN repeat-containing protein 3-like [Lingula anatina] 4e-54; gi|405976919|gb|EKC41397.1|MORN repeat-containing protein 3 [Crassostrea gigas] 5e-54; | ||||||
| NvERTx.4.108337 | R-5 | 0.586074534631 | E-5 | 0.99999999985 | E7FAM5 | E3 ubiquitin-protein ligase TRIM71 | gi|156370888|ref|XP_001628499.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999980663|gb|KXJ19837.1|E3 ubiquitin-protein ligase TRIM71 [Exaiptasia pallida] 0e+00; gi|1005458311|ref|XP_015766287.1|PREDICTED: E3 ubiquitin-protein ligase TRIM71-like [Acropora digitifera] 9e-291; gi|999980696|gb|KXJ19870.1|E3 ubiquitin-protein ligase TRIM71 [Exaiptasia pallida] 8e-159; gi|156370856|ref|XP_001628483.1|predicted protein [Nematostella vectensis] 4e-137; gi|1005458194|ref|XP_015766225.1|PREDICTED: E3 ubiquitin-protein ligase TRIM71-like, partial [Acropora digitifera] 5e-119; gi|1005439105|ref|XP_015756960.1|PREDICTED: E3 ubiquitin-protein ligase TRIM71-like [Acropora digitifera] 1e-113; gi|1005471833|ref|XP_015772779.1|PREDICTED: E3 ubiquitin-protein ligase TRIM71-like [Acropora digitifera] 3e-108; gi|1005484892|ref|XP_015779062.1|PREDICTED: E3 ubiquitin-protein ligase TRIM71-like isoform X1 [Acropora digitifera] 1e-103; gi|1005484894|ref|XP_015779063.1|PREDICTED: E3 ubiquitin-protein ligase TRIM71-like isoform X2 [Acropora digitifera] 2e-103; | ||||||
| NvERTx.4.165642 | R-7 | 0.524036613322 | E-5 | 0.999999999846 | P59999 | Actin-related protein 2/3 complex subunit 4 | gi|156398184|ref|XP_001638069.1|predicted protein [Nematostella vectensis], 4e-87 |
| other nr_hits | gi|999978542|gb|KXJ17716.1|Actin-related protein 2/3 complex subunit 4 [Exaiptasia pallida] 4e-83; gi|1005465437|ref|XP_015769713.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Acropora digitifera] 1e-82; gi|340368165|ref|XP_003382623.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Amphimedon queenslandica] 3e-81; gi|1007748435|ref|XP_015813949.1|PREDICTED: actin-related protein 2/3 complex subunit 4-like [Nothobranchius furzeri] 5e-80; gi|928032090|ref|XP_013864527.1|PREDICTED: actin-related protein 2/3 complex subunit 4-like [Austrofundulus limnaeus] 7e-80; gi|637273547|ref|XP_008103770.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Anolis carolinensis] 7e-80; gi|119640029|gb|ABL85456.1|actin related protein 2/3 complex subunit 4 [Suberites domuncula] 7e-80; gi|1126171228|ref|XP_019617647.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Branchiostoma belcheri] 9e-80; gi|1101536967|ref|XP_018934293.1|PREDICTED: actin-related protein 2/3 complex subunit 4-like isoform X1 [Cyprinus carpio] 1e-79; | ||||||
| NvERTx.4.165643 | R-7 | 0.524036613322 | E-5 | 0.999999999846 | P59999 | Actin-related protein 2/3 complex subunit 4 | gi|156398184|ref|XP_001638069.1|predicted protein [Nematostella vectensis], 4e-87 |
| other nr_hits | gi|999978542|gb|KXJ17716.1|Actin-related protein 2/3 complex subunit 4 [Exaiptasia pallida] 4e-83; gi|1005465437|ref|XP_015769713.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Acropora digitifera] 1e-82; gi|340368165|ref|XP_003382623.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Amphimedon queenslandica] 3e-81; gi|1007748435|ref|XP_015813949.1|PREDICTED: actin-related protein 2/3 complex subunit 4-like [Nothobranchius furzeri] 5e-80; gi|928032090|ref|XP_013864527.1|PREDICTED: actin-related protein 2/3 complex subunit 4-like [Austrofundulus limnaeus] 7e-80; gi|637273547|ref|XP_008103770.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Anolis carolinensis] 7e-80; gi|119640029|gb|ABL85456.1|actin related protein 2/3 complex subunit 4 [Suberites domuncula] 7e-80; gi|1126171228|ref|XP_019617647.1|PREDICTED: actin-related protein 2/3 complex subunit 4 [Branchiostoma belcheri] 9e-80; gi|1101536967|ref|XP_018934293.1|PREDICTED: actin-related protein 2/3 complex subunit 4-like isoform X1 [Cyprinus carpio] 1e-79; | ||||||
| NvERTx.4.41110 | R-7 | 0.890690384009 | E-5 | 0.999999999846 | I3IKN2_9BACT | Uncharacterized protein {ECO:0000313|EMBL:GAB62277.1} | gi|386404441|dbj|GAB62277.1|conserved hypothetical protein [Candidatus Jettenia caeni], 1e-08 |
| other nr_hits | gi|553740096|ref|WP_023074280.1|cupin domain-containing protein [Leptolyngbya sp. Heron Island J] 4e-08; gi|299471185|embl|CBN79041.1|conserved unknown protein [Ectocarpus siliculosus] 5e-08; gi|145340817|ref|XP_001415514.1|predicted protein [Ostreococcus lucimarinus CCE9901] 1e-07; gi|612386767|ref|XP_007508905.1|COG1917: Uncharacterized conserved protein, contains double-stranded beta-helix domain (ISS) [Bathycoccus prasinos] 2e-07; gi|493565646|ref|WP_006518988.1|cupin domain-containing protein [Leptolyngbya sp. PCC 7375] 6e-06; gi|308812320|ref|XP_003083467.1|COG1917: Uncharacterized conserved protein, contains double-stranded beta-helix domain (ISS) [Ostreococcus tauri] 8e-06; gi|1094599647|embl|SEL86435.1|Cupin domain-containing protein [Maribacter orientalis] 1e-05; gi|740239935|ref|WP_038081025.1|cupin [Tolypothrix bouteillei] 1e-05; gi|504963270|ref|WP_015150372.1|cupin domain-containing protein [Oscillatoria acuminata] 3e-05; | ||||||
| NvERTx.4.136584 | R-7 | 0.890690384009 | E-5 | 0.999999999846 | O35218 | Cleavage and polyadenylation specificity factor subunit 2 | gi|156399337|ref|XP_001638458.1|predicted protein [Nematostella vectensis], 1e-259 |
| other nr_hits | gi|999982919|gb|KXJ22065.1|Cleavage and polyadenylation specificity factor subunit 2 [Exaiptasia pallida] 4e-230; gi|828189945|ref|XP_004205466.2|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Hydra vulgaris] 8e-177; gi|1024946651|ref|XP_016362391.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Sinocyclocheilus anshuiensis] 2e-174; gi|1049225873|ref|XP_017551173.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Pygocentrus nattereri] 5e-174; gi|390333491|ref|XP_780045.3|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Strongylocentrotus purpuratus] 5e-174; gi|919096261|ref|XP_013384817.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Lingula anatina] 7e-174; gi|1123925526|ref|XP_019600859.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Rhinolophus sinicus] 9e-174; gi|573889964|ref|XP_006632745.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Lepisosteus oculatus] 9e-174; gi|1040673596|ref|XP_017207953.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Danio rerio] 1e-173; | ||||||
| NvERTx.4.136585 | R-7 | 0.890690384009 | E-5 | 0.999999999846 | Q9P2I0 | Cleavage and polyadenylation specificity factor subunit 2 | gi|156399337|ref|XP_001638458.1|predicted protein [Nematostella vectensis], 4e-162 |
| other nr_hits | gi|999982919|gb|KXJ22065.1|Cleavage and polyadenylation specificity factor subunit 2 [Exaiptasia pallida] 6e-148; gi|390333491|ref|XP_780045.3|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Strongylocentrotus purpuratus] 3e-122; gi|828189945|ref|XP_004205466.2|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Hydra vulgaris] 7e-122; gi|676479093|ref|XP_009061636.1|hypothetical protein LOTGIDRAFT_219936 [Lottia gigantea] 7e-122; gi|919096261|ref|XP_013384817.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Lingula anatina] 1e-121; gi|1139845377|ref|XP_011415147.2|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Crassostrea gigas] 2e-120; gi|585682229|ref|XP_006812203.1|PREDICTED: LOW QUALITY PROTEIN: cleavage and polyadenylation specificity factor subunit 2-like [Saccoglossus kowalevskii] 4e-120; gi|926623469|ref|XP_013777155.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Limulus polyphemus] 5e-120; gi|675365290|gb|KFM58192.1|Cleavage and polyadenylation specificity factor subunit 2, partial [Stegodyphus mimosarum] 5e-120; | ||||||
| NvERTx.4.136586 | R-7 | 0.890690384009 | E-5 | 0.999999999846 | O35218 | Cleavage and polyadenylation specificity factor subunit 2 | gi|156399337|ref|XP_001638458.1|predicted protein [Nematostella vectensis], 2e-297 |
| other nr_hits | gi|999982919|gb|KXJ22065.1|Cleavage and polyadenylation specificity factor subunit 2 [Exaiptasia pallida] 9e-265; gi|828189945|ref|XP_004205466.2|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Hydra vulgaris] 2e-197; gi|919096261|ref|XP_013384817.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Lingula anatina] 4e-197; gi|1139845377|ref|XP_011415147.2|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Crassostrea gigas] 7e-195; gi|585682229|ref|XP_006812203.1|PREDICTED: LOW QUALITY PROTEIN: cleavage and polyadenylation specificity factor subunit 2-like [Saccoglossus kowalevskii] 8e-194; gi|390333491|ref|XP_780045.3|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Strongylocentrotus purpuratus] 8e-194; gi|675365290|gb|KFM58192.1|Cleavage and polyadenylation specificity factor subunit 2, partial [Stegodyphus mimosarum] 1e-192; gi|1024946651|ref|XP_016362391.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Sinocyclocheilus anshuiensis] 3e-192; gi|573889964|ref|XP_006632745.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Lepisosteus oculatus] 3e-192; | ||||||
| NvERTx.4.136587 | R-7 | 0.890690384009 | E-5 | 0.999999999846 | O35218 | Cleavage and polyadenylation specificity factor subunit 2 | gi|156399337|ref|XP_001638458.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999982919|gb|KXJ22065.1|Cleavage and polyadenylation specificity factor subunit 2 [Exaiptasia pallida] 0e+00; gi|828189945|ref|XP_004205466.2|PREDICTED: cleavage and polyadenylation specificity factor subunit 2-like [Hydra vulgaris] 3e-249; gi|632971170|ref|XP_007902040.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Callorhinchus milii] 5e-243; gi|573889964|ref|XP_006632745.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Lepisosteus oculatus] 1e-242; gi|556947150|ref|XP_005986658.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Latimeria chalumnae] 3e-242; gi|1016670379|ref|XP_016061204.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Miniopterus natalensis] 2e-241; gi|1123925526|ref|XP_019600859.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Rhinolophus sinicus] 4e-241; gi|1041089704|ref|XP_017267215.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 [Kryptolebias marmoratus] 6e-241; gi|602631389|ref|XP_007422598.1|PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Python bivittatus] 6e-241; | ||||||
