Explore Mfuzz Clusters
Mfuzz cluster
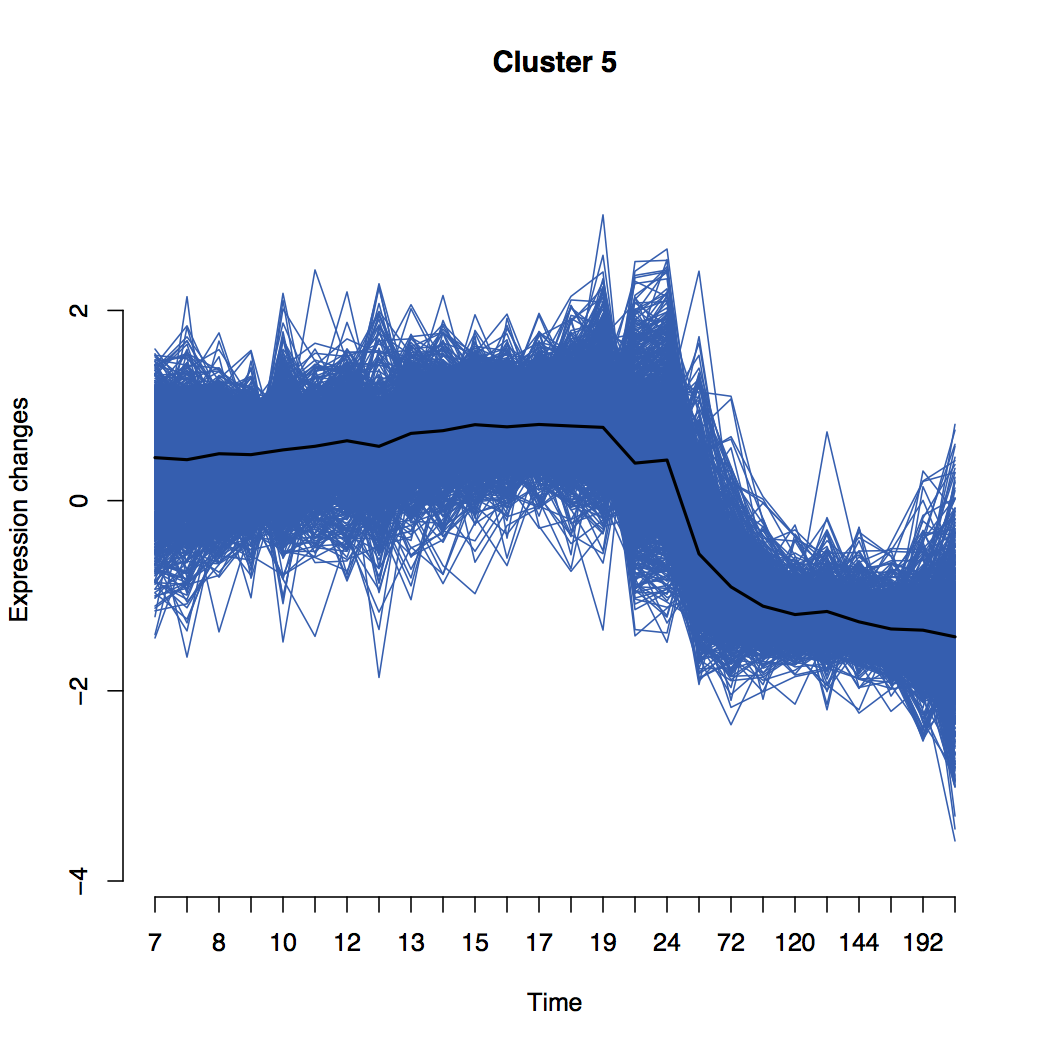
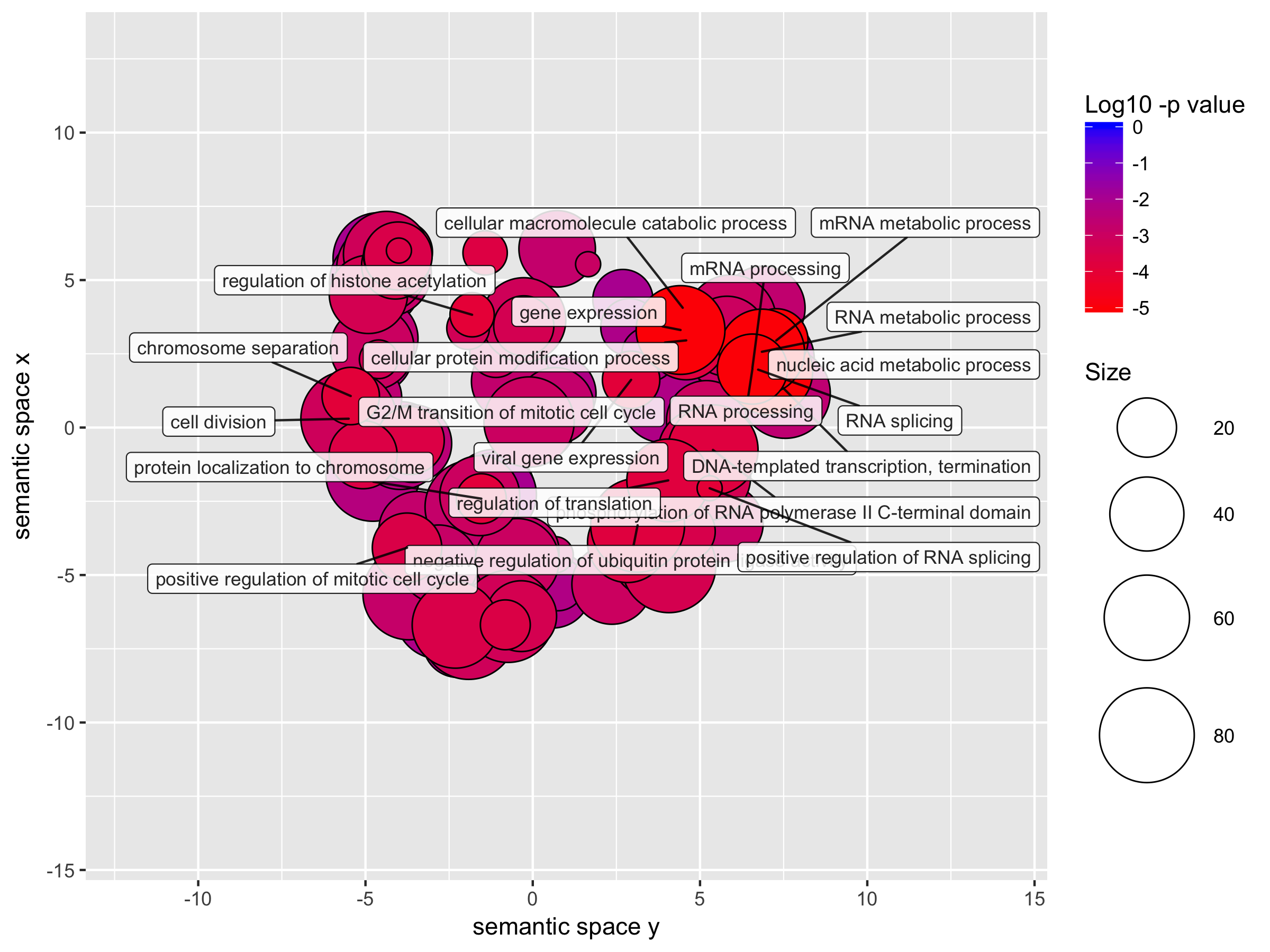
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.130514 | R-8 | 0.886041113654 | E-5 | 0.999999999964 | P02545 | Prelamin-A/C | gi|399145793|gb|AFP25101.1|cytovec [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|1005477654|ref|XP_015775595.1|PREDICTED: prelamin-A/C-like [Acropora digitifera] 2e-209; gi|999971639|gb|KXJ10813.1|Lamin tail domain-containing protein 1 [Exaiptasia pallida] 3e-134; gi|1126197526|ref|XP_019631946.1|PREDICTED: lamin-A-like isoform X1 [Branchiostoma belcheri] 9e-109; gi|291232460|ref|XP_002736176.1|PREDICTED: prelamin-A/C-like [Saccoglossus kowalevskii] 7e-106; gi|1126197528|ref|XP_019631948.1|PREDICTED: lamin-B1-like isoform X2 [Branchiostoma belcheri] 4e-105; gi|780146506|ref|XP_011681426.1|PREDICTED: lamin-B2 [Strongylocentrotus purpuratus] 2e-103; gi|1126197530|ref|XP_019631949.1|PREDICTED: lamin-A-like isoform X3 [Branchiostoma belcheri] 3e-103; gi|190014566|dbj|BAG48261.1|nematocilin A [Hydra vulgaris] 8e-68; gi|449686858|ref|XP_002166919.2|PREDICTED: lamin-C [Hydra vulgaris] 5e-67; | ||||||
| NvERTx.4.114587 | R-7 | 0.999003953859 | E-5 | 0.999999999961 | Q8HXM1 | Cleavage stimulation factor subunit 2 | gi|999985249|gb|KXJ24390.1|Cleavage stimulation factor subunit 2 [Exaiptasia pallida], 2e-94 |
| other nr_hits | gi|1126211029|ref|XP_019639290.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X2 [Branchiostoma belcheri] 2e-80; gi|1126211033|ref|XP_019639293.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X4 [Branchiostoma belcheri] 2e-80; gi|1126211031|ref|XP_019639292.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X3 [Branchiostoma belcheri] 2e-80; gi|1126211027|ref|XP_019639289.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X1 [Branchiostoma belcheri] 2e-80; gi|926637712|ref|XP_013784845.1|PREDICTED: cleavage stimulation factor subunit 2-like [Limulus polyphemus] 1e-79; gi|1072281532|ref|XP_018426950.1|PREDICTED: cleavage stimulation factor subunit 2 [Nanorana parkeri] 5e-79; gi|632500|gb|AAB50269.1|polyadenylation factor 64 kDa subunit [Xenopus laevis] 1e-78; gi|847153465|ref|XP_012823534.1|PREDICTED: cleavage stimulation factor subunit 2 isoform X1 [Xenopus tropicalis] 1e-78; gi|847153467|ref|XP_012823536.1|PREDICTED: cleavage stimulation factor subunit 2 isoform X2 [Xenopus tropicalis] 1e-78; | ||||||
| NvERTx.4.114588 | R-7 | 0.999003953859 | E-5 | 0.999999999961 | Q8HXM1 | Cleavage stimulation factor subunit 2 | gi|999985249|gb|KXJ24390.1|Cleavage stimulation factor subunit 2 [Exaiptasia pallida], 2e-94 |
| other nr_hits | gi|1126211029|ref|XP_019639290.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X2 [Branchiostoma belcheri] 2e-80; gi|1126211033|ref|XP_019639293.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X4 [Branchiostoma belcheri] 2e-80; gi|1126211031|ref|XP_019639292.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X3 [Branchiostoma belcheri] 2e-80; gi|1126211027|ref|XP_019639289.1|PREDICTED: cleavage stimulation factor subunit 2-like isoform X1 [Branchiostoma belcheri] 2e-80; gi|926637712|ref|XP_013784845.1|PREDICTED: cleavage stimulation factor subunit 2-like [Limulus polyphemus] 1e-79; gi|1072281532|ref|XP_018426950.1|PREDICTED: cleavage stimulation factor subunit 2 [Nanorana parkeri] 5e-79; gi|632500|gb|AAB50269.1|polyadenylation factor 64 kDa subunit [Xenopus laevis] 1e-78; gi|847153465|ref|XP_012823534.1|PREDICTED: cleavage stimulation factor subunit 2 isoform X1 [Xenopus tropicalis] 1e-78; gi|847153467|ref|XP_012823536.1|PREDICTED: cleavage stimulation factor subunit 2 isoform X2 [Xenopus tropicalis] 1e-78; | ||||||
| NvERTx.4.51076 | R-7 | 0.996261931785 | E-5 | 0.99999999996 | Q9WVA3 | Mitotic checkpoint protein BUB3 | gi|156390723|ref|XP_001635419.1|predicted protein [Nematostella vectensis], 2e-93 |
| other nr_hits | gi|999990563|gb|KXJ29585.1|Mitotic checkpoint protein BUB3 [Exaiptasia pallida] 3e-84; gi|1005442560|ref|XP_015758620.1|PREDICTED: mitotic checkpoint protein BUB3-like [Acropora digitifera] 4e-80; gi|1005479802|ref|XP_015776631.1|PREDICTED: mitotic checkpoint protein BUB3-like [Acropora digitifera] 4e-80; gi|585639434|ref|XP_006879938.1|PREDICTED: mitotic checkpoint protein BUB3 [Elephantulus edwardii] 5e-80; gi|260826716|ref|XP_002608311.1|hypothetical protein BRAFLDRAFT_125481 [Branchiostoma floridae] 5e-80; gi|1126223928|ref|XP_019646291.1|PREDICTED: mitotic checkpoint protein BUB3-like [Branchiostoma belcheri] 1e-79; gi|852765607|ref|XP_012877401.1|PREDICTED: mitotic checkpoint protein BUB3 isoform X1 [Dipodomys ordii] 1e-79; gi|852765610|ref|XP_012877403.1|PREDICTED: mitotic checkpoint protein BUB3 isoform X2 [Dipodomys ordii] 1e-79; gi|1147351072|ref|XP_020027367.1|mitotic checkpoint protein BUB3 isoform X2 [Castor canadensis] 1e-79; | ||||||
| NvERTx.4.51077 | R-7 | 0.996261931785 | E-5 | 0.99999999996 | Q9WVA3 | Mitotic checkpoint protein BUB3 | gi|156390723|ref|XP_001635419.1|predicted protein [Nematostella vectensis], 4e-181 |
| other nr_hits | gi|999990563|gb|KXJ29585.1|Mitotic checkpoint protein BUB3 [Exaiptasia pallida] 2e-147; gi|1005442560|ref|XP_015758620.1|PREDICTED: mitotic checkpoint protein BUB3-like [Acropora digitifera] 4e-140; gi|260826716|ref|XP_002608311.1|hypothetical protein BRAFLDRAFT_125481 [Branchiostoma floridae] 3e-134; gi|1126223928|ref|XP_019646291.1|PREDICTED: mitotic checkpoint protein BUB3-like [Branchiostoma belcheri] 3e-133; gi|1121911950|ref|XP_019524701.1|PREDICTED: mitotic checkpoint protein BUB3 [Hipposideros armiger] 6e-133; gi|852765607|ref|XP_012877401.1|PREDICTED: mitotic checkpoint protein BUB3 isoform X1 [Dipodomys ordii] 1e-132; gi|852765610|ref|XP_012877403.1|PREDICTED: mitotic checkpoint protein BUB3 isoform X2 [Dipodomys ordii] 1e-132; gi|507551046|ref|XP_004659326.1|PREDICTED: mitotic checkpoint protein BUB3 [Jaculus jaculus] 1e-132; gi|1147351072|ref|XP_020027367.1|mitotic checkpoint protein BUB3 isoform X2 [Castor canadensis] 1e-132; | ||||||
| NvERTx.4.68920 | R-6 | 0.999128306169 | E-5 | 0.999999999959 | C3ZC99_BRAFL | Putative uncharacterized protein {ECO:0000313|EMBL:EEN49871.1} | gi|156388917|ref|XP_001634739.1|predicted protein [Nematostella vectensis], 1e-112 |
| other nr_hits | gi|999984546|gb|KXJ23687.1|MANSC domain-containing protein 1 [Exaiptasia pallida] 4e-35; gi|390354732|ref|XP_003728395.1|PREDICTED: uncharacterized protein LOC100891046 isoform X2 [Strongylocentrotus purpuratus] 9e-13; gi|260797741|ref|XP_002593860.1|hypothetical protein BRAFLDRAFT_75680 [Branchiostoma floridae] 4e-09; gi|585676116|ref|XP_006818898.1|PREDICTED: uncharacterized protein C11orf24-like isoform X1 [Saccoglossus kowalevskii] 2e-08; gi|585676120|ref|XP_006818899.1|PREDICTED: uncharacterized protein C11orf24-like isoform X2 [Saccoglossus kowalevskii] 2e-08; gi|260797769|ref|XP_002593874.1|hypothetical protein BRAFLDRAFT_75668 [Branchiostoma floridae] 2e-08; gi|1126208924|ref|XP_019638149.1|PREDICTED: salivary glue protein Sgs-3-like [Branchiostoma belcheri] 4e-07; gi|390354730|ref|XP_003728394.1|PREDICTED: proline-rich receptor-like protein kinase PERK8 isoform X1 [Strongylocentrotus purpuratus] 3e-06; gi|340377413|ref|XP_003387224.1|PREDICTED: uncharacterized protein C11orf24 homolog [Amphimedon queenslandica] 1e-05; | ||||||
| NvERTx.4.138494 | R-6 | 0.998315813895 | E-5 | 0.999999999958 | Q96JK2 | DDB1- and CUL4-associated factor 5 | gi|999978565|gb|KXJ17739.1|DDB1- and CUL4-associated factor 5 [Exaiptasia pallida], 3e-130 |
| other nr_hits | gi|1005435672|ref|XP_015755345.1|PREDICTED: DDB1- and CUL4-associated factor 5-like [Acropora digitifera] 4e-116; gi|762124110|ref|XP_011445058.1|PREDICTED: DDB1- and CUL4-associated factor 5-like [Crassostrea gigas] 3e-103; gi|742204810|ref|XP_010894570.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X1 [Esox lucius] 9e-103; gi|742204814|ref|XP_010894571.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X2 [Esox lucius] 9e-103; gi|1126226492|ref|XP_019647677.1|PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC109487982 [Branchiostoma belcheri] 3e-102; gi|1072269830|ref|XP_018420602.1|PREDICTED: DDB1- and CUL4-associated factor 5 [Nanorana parkeri] 3e-102; gi|975106782|ref|XP_015268801.1|PREDICTED: DDB1- and CUL4-associated factor 5 [Gekko japonicus] 4e-102; gi|410897855|ref|XP_003962414.1|PREDICTED: DDB1- and CUL4-associated factor 5 [Takifugu rubripes] 4e-102; gi|1083395123|ref|XP_018582946.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X2 [Scleropages formosus] 1e-101; | ||||||
| NvERTx.4.138496 | R-6 | 0.998315813895 | E-5 | 0.999999999958 | Q80T85 | DDB1- and CUL4-associated factor 5 | gi|999978565|gb|KXJ17739.1|DDB1- and CUL4-associated factor 5 [Exaiptasia pallida], 2e-87 |
| other nr_hits | gi|156381976|ref|XP_001632331.1|predicted protein [Nematostella vectensis] 2e-64; gi|1005435672|ref|XP_015755345.1|PREDICTED: DDB1- and CUL4-associated factor 5-like [Acropora digitifera] 2e-61; gi|918999032|ref|XP_013397984.1|PREDICTED: DDB1- and CUL4-associated factor 5-like [Lingula anatina] 2e-59; gi|919016711|ref|XP_013392731.1|PREDICTED: uncharacterized protein LOC106160626, partial [Lingula anatina] 3e-59; gi|1126226492|ref|XP_019647677.1|PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC109487982 [Branchiostoma belcheri] 7e-58; gi|803096080|ref|XP_012037291.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X1 [Ovis aries] 7e-58; gi|426233580|ref|XP_004010794.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X2 [Ovis aries] 7e-58; gi|585655027|ref|XP_006903754.1|PREDICTED: LOW QUALITY PROTEIN: DDB1 and CUL4 associated factor 5 [Elephantulus edwardii] 2e-57; gi|815820372|ref|XP_012231404.1|PREDICTED: DDB1- and CUL4-associated factor 5 [Linepithema humile] 2e-57; | ||||||
| NvERTx.4.138497 | R-6 | 0.998315813895 | E-5 | 0.999999999958 | Q96JK2 | DDB1- and CUL4-associated factor 5 | gi|999978565|gb|KXJ17739.1|DDB1- and CUL4-associated factor 5 [Exaiptasia pallida], 5e-170 |
| other nr_hits | gi|1005435672|ref|XP_015755345.1|PREDICTED: DDB1- and CUL4-associated factor 5-like [Acropora digitifera] 5e-137; gi|975106782|ref|XP_015268801.1|PREDICTED: DDB1- and CUL4-associated factor 5 [Gekko japonicus] 7e-124; gi|762124110|ref|XP_011445058.1|PREDICTED: DDB1- and CUL4-associated factor 5-like [Crassostrea gigas] 7e-124; gi|410897855|ref|XP_003962414.1|PREDICTED: DDB1- and CUL4-associated factor 5 [Takifugu rubripes] 9e-124; gi|1126226492|ref|XP_019647677.1|PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC109487982 [Branchiostoma belcheri] 3e-123; gi|426233580|ref|XP_004010794.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X2 [Ovis aries] 1e-122; gi|927130938|ref|XP_013909888.1|PREDICTED: DDB1- and CUL4-associated factor 5 [Thamnophis sirtalis] 2e-122; gi|742204810|ref|XP_010894570.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X1 [Esox lucius] 2e-122; gi|658914738|ref|XP_008397193.1|PREDICTED: DDB1- and CUL4-associated factor 5 isoform X1 [Poecilia reticulata] 2e-122; | ||||||
| NvERTx.4.183870 | R-2 | 0.967028063322 | E-5 | 0.999999999957 | Q9R1R2 | Tripartite motif-containing protein 3 | gi|156384131|ref|XP_001633185.1|predicted protein [Nematostella vectensis], 7e-84 |
| other nr_hits | gi|999972222|gb|KXJ11396.1|hypothetical protein AC249_AIPGENE8042 [Exaiptasia pallida] 3e-62; gi|1005445382|ref|XP_015759994.1|PREDICTED: tripartite motif-containing protein 67-like [Acropora digitifera] 1e-55; gi|973182799|ref|XP_015215459.1|PREDICTED: E3 ubiquitin-protein ligase TRIM56 [Lepisosteus oculatus] 6e-11; gi|919070152|ref|XP_013416573.1|PREDICTED: RING finger protein 17-like [Lingula anatina] 2e-10; gi|632982189|ref|XP_007908000.1|PREDICTED: uncharacterized protein LOC103189419 [Callorhinchus milii] 1e-09; gi|678320380|embl|CDW84267.1|kelch motif family protein [Stylonychia lemnae] 5e-09; gi|115727664|ref|XP_001199620.1|PREDICTED: tripartite motif-containing protein 2-like [Strongylocentrotus purpuratus] 5e-09; gi|1126211117|ref|XP_019639337.1|PREDICTED: transcription elongation factor SPT5-like [Branchiostoma belcheri] 7e-09; gi|260808476|ref|XP_002599033.1|hypothetical protein BRAFLDRAFT_104271 [Branchiostoma floridae] 7e-09; | ||||||
| NvERTx.4.183871 | R-2 | 0.967028063322 | E-5 | 0.999999999957 | Q9R1R2 | Tripartite motif-containing protein 3 | gi|156384131|ref|XP_001633185.1|predicted protein [Nematostella vectensis], 7e-84 |
| other nr_hits | gi|999972222|gb|KXJ11396.1|hypothetical protein AC249_AIPGENE8042 [Exaiptasia pallida] 3e-62; gi|1005445382|ref|XP_015759994.1|PREDICTED: tripartite motif-containing protein 67-like [Acropora digitifera] 1e-55; gi|973182799|ref|XP_015215459.1|PREDICTED: E3 ubiquitin-protein ligase TRIM56 [Lepisosteus oculatus] 6e-11; gi|919070152|ref|XP_013416573.1|PREDICTED: RING finger protein 17-like [Lingula anatina] 2e-10; gi|632982189|ref|XP_007908000.1|PREDICTED: uncharacterized protein LOC103189419 [Callorhinchus milii] 1e-09; gi|678320380|embl|CDW84267.1|kelch motif family protein [Stylonychia lemnae] 5e-09; gi|115727664|ref|XP_001199620.1|PREDICTED: tripartite motif-containing protein 2-like [Strongylocentrotus purpuratus] 5e-09; gi|1126211117|ref|XP_019639337.1|PREDICTED: transcription elongation factor SPT5-like [Branchiostoma belcheri] 7e-09; gi|260808476|ref|XP_002599033.1|hypothetical protein BRAFLDRAFT_104271 [Branchiostoma floridae] 7e-09; | ||||||
| NvERTx.4.183874 | R-2 | 0.967028063322 | E-5 | 0.999999999957 | Q9UPN9 | E3 ubiquitin-protein ligase TRIM33 | gi|156384131|ref|XP_001633185.1|predicted protein [Nematostella vectensis], 2e-170 |
| other nr_hits | gi|1005445382|ref|XP_015759994.1|PREDICTED: tripartite motif-containing protein 67-like [Acropora digitifera] 6e-95; gi|999972222|gb|KXJ11396.1|hypothetical protein AC249_AIPGENE8042 [Exaiptasia pallida] 3e-20; gi|1133460172|ref|XP_019851256.1|PREDICTED: probable E3 ubiquitin-protein ligase MID2 isoform X2 [Amphimedon queenslandica] 2e-12; gi|761908050|ref|XP_011403547.1|PREDICTED: probable E3 ubiquitin-protein ligase MID2 isoform X1 [Amphimedon queenslandica] 2e-12; gi|973209284|ref|XP_015224085.1|PREDICTED: E3 ubiquitin/ISG15 ligase TRIM25-like [Lepisosteus oculatus] 6e-08; gi|1126225727|ref|XP_019647277.1|PREDICTED: uncharacterized protein LOC109487691 isoform X1 [Branchiostoma belcheri] 2e-07; gi|1126225729|ref|XP_019647278.1|PREDICTED: uncharacterized protein LOC109487691 isoform X2 [Branchiostoma belcheri] 2e-07; gi|260833066|ref|XP_002611478.1|hypothetical protein BRAFLDRAFT_63887 [Branchiostoma floridae] 2e-07; gi|926616855|ref|XP_013773565.1|PREDICTED: E3 ubiquitin-protein ligase TRIM9-like [Limulus polyphemus] 5e-07; | ||||||
| NvERTx.4.183875 | R-2 | 0.967028063322 | E-5 | 0.999999999957 | Q9UPN9 | E3 ubiquitin-protein ligase TRIM33 | gi|156384131|ref|XP_001633185.1|predicted protein [Nematostella vectensis], 2e-170 |
| other nr_hits | gi|1005445382|ref|XP_015759994.1|PREDICTED: tripartite motif-containing protein 67-like [Acropora digitifera] 6e-95; gi|999972222|gb|KXJ11396.1|hypothetical protein AC249_AIPGENE8042 [Exaiptasia pallida] 3e-20; gi|1133460172|ref|XP_019851256.1|PREDICTED: probable E3 ubiquitin-protein ligase MID2 isoform X2 [Amphimedon queenslandica] 2e-12; gi|761908050|ref|XP_011403547.1|PREDICTED: probable E3 ubiquitin-protein ligase MID2 isoform X1 [Amphimedon queenslandica] 2e-12; gi|973209284|ref|XP_015224085.1|PREDICTED: E3 ubiquitin/ISG15 ligase TRIM25-like [Lepisosteus oculatus] 6e-08; gi|1126225727|ref|XP_019647277.1|PREDICTED: uncharacterized protein LOC109487691 isoform X1 [Branchiostoma belcheri] 2e-07; gi|1126225729|ref|XP_019647278.1|PREDICTED: uncharacterized protein LOC109487691 isoform X2 [Branchiostoma belcheri] 2e-07; gi|260833066|ref|XP_002611478.1|hypothetical protein BRAFLDRAFT_63887 [Branchiostoma floridae] 2e-07; gi|926616855|ref|XP_013773565.1|PREDICTED: E3 ubiquitin-protein ligase TRIM9-like [Limulus polyphemus] 5e-07; | ||||||
| NvERTx.4.35311 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.126583 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | D2HNY3 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|999979851|gb|KXJ19025.1|Fanconi-associated nuclease 1 [Exaiptasia pallida], 1e-15 |
| other nr_hits | gi|149410836|ref|XP_001509834.1|PREDICTED: fanconi-associated nuclease 1 [Ornithorhynchus anatinus] 1e-13; gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera] 1e-13; gi|410908619|ref|XP_003967788.1|PREDICTED: fanconi-associated nuclease 1 [Takifugu rubripes] 2e-13; gi|47220527|embl|CAG05553.1|unnamed protein product, partial [Tetraodon nigroviridis] 3e-13; gi|677411172|gb|KFQ19035.1|Fanconi-associated nuclease 1, partial [Merops nubicus] 4e-13; gi|675624464|ref|XP_008940462.1|PREDICTED: fanconi-associated nuclease 1 [Merops nubicus] 4e-13; gi|556950934|ref|XP_005987805.1|PREDICTED: fanconi-associated nuclease 1 [Latimeria chalumnae] 4e-13; gi|975119100|ref|XP_015274978.1|PREDICTED: fanconi-associated nuclease 1 [Gekko japonicus] 7e-13; gi|1143361521|ref|XP_019969572.1|PREDICTED: fanconi-associated nuclease 1 [Paralichthys olivaceus] 9e-13; | ||||||
| NvERTx.4.126595 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | Q1LWH4 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera], 1e-59 |
| other nr_hits | gi|999979843|gb|KXJ19017.1|Fanconi-associated nuclease 1 [Exaiptasia pallida] 6e-47; gi|291232927|ref|XP_002736405.1|PREDICTED: fanconi-associated nuclease 1-like [Saccoglossus kowalevskii] 6e-42; gi|1126188815|ref|XP_019627245.1|PREDICTED: fanconi-associated nuclease 1-like [Branchiostoma belcheri] 2e-41; gi|1025381900|ref|XP_016400729.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus rhinocerous] 3e-39; gi|1025003865|ref|XP_016320960.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Sinocyclocheilus anshuiensis] 1e-38; gi|1101627130|ref|XP_018977325.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Cyprinus carpio] 1e-38; gi|1101627132|ref|XP_018977326.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Cyprinus carpio] 1e-38; gi|1025003863|ref|XP_016320959.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus anshuiensis] 1e-38; gi|160409999|sp|FAN1_DANRE|Q1LWH4.2|RecName: Full=Fanconi-associated nuclease 1; AltName: Full=FANCD2/FANCI-associated nuclease 1; AltName: Full=Myotubularin-related protein 15 2e-38; | ||||||
| NvERTx.4.126596 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | Q69ZT1 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|999979851|gb|KXJ19025.1|Fanconi-associated nuclease 1 [Exaiptasia pallida], 1e-91 |
| other nr_hits | gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera] 5e-81; gi|1005453634|ref|XP_015763977.1|PREDICTED: fanconi-associated nuclease 1-like [Acropora digitifera] 5e-71; gi|704308333|ref|XP_010165142.1|PREDICTED: fanconi-associated nuclease 1 [Antrostomus carolinensis] 8e-59; gi|327284500|ref|XP_003226975.1|PREDICTED: fanconi-associated nuclease 1 [Anolis carolinensis] 1e-58; gi|726997619|ref|XP_010389092.1|PREDICTED: fanconi-associated nuclease 1 [Corvus cornix cornix] 2e-58; gi|696965341|ref|XP_009553624.1|PREDICTED: fanconi-associated nuclease 1 [Cuculus canorus] 2e-58; gi|676582200|gb|KFO72063.1|Fanconi-associated nuclease 1, partial [Cuculus canorus] 2e-58; gi|698473658|ref|XP_009706834.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like, partial [Cariama cristata] 3e-58; gi|676408976|gb|KFO53385.1|Fanconi-associated nuclease 1, partial [Corvus brachyrhynchos] 3e-58; | ||||||
| NvERTx.4.126597 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | Q1LWH4 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera], 1e-59 |
| other nr_hits | gi|999979843|gb|KXJ19017.1|Fanconi-associated nuclease 1 [Exaiptasia pallida] 6e-47; gi|291232927|ref|XP_002736405.1|PREDICTED: fanconi-associated nuclease 1-like [Saccoglossus kowalevskii] 6e-42; gi|1126188815|ref|XP_019627245.1|PREDICTED: fanconi-associated nuclease 1-like [Branchiostoma belcheri] 2e-41; gi|1025381900|ref|XP_016400729.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus rhinocerous] 3e-39; gi|1025003865|ref|XP_016320960.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Sinocyclocheilus anshuiensis] 1e-38; gi|1101627130|ref|XP_018977325.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Cyprinus carpio] 1e-38; gi|1101627132|ref|XP_018977326.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Cyprinus carpio] 1e-38; gi|1025003863|ref|XP_016320959.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus anshuiensis] 1e-38; gi|160409999|sp|FAN1_DANRE|Q1LWH4.2|RecName: Full=Fanconi-associated nuclease 1; AltName: Full=FANCD2/FANCI-associated nuclease 1; AltName: Full=Myotubularin-related protein 15 2e-38; | ||||||
| NvERTx.4.126598 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | Q1LWH4 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera], 1e-212 |
| other nr_hits | gi|999979851|gb|KXJ19025.1|Fanconi-associated nuclease 1 [Exaiptasia pallida] 3e-174; gi|1126188815|ref|XP_019627245.1|PREDICTED: fanconi-associated nuclease 1-like [Branchiostoma belcheri] 4e-154; gi|1049241537|ref|XP_017559706.1|PREDICTED: fanconi-associated nuclease 1 [Pygocentrus nattereri] 3e-151; gi|530582424|ref|XP_005285058.1|PREDICTED: fanconi-associated nuclease 1 isoform X1 [Chrysemys picta bellii] 1e-150; gi|1079793502|ref|XP_018521936.1|PREDICTED: fanconi-associated nuclease 1 [Lates calcarifer] 1e-149; gi|564230821|ref|XP_006259814.1|PREDICTED: fanconi-associated nuclease 1 isoform X1 [Alligator mississippiensis] 1e-149; gi|410908619|ref|XP_003967788.1|PREDICTED: fanconi-associated nuclease 1 [Takifugu rubripes] 1e-149; gi|1101627130|ref|XP_018977325.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Cyprinus carpio] 1e-149; gi|736245438|ref|XP_010786339.1|PREDICTED: fanconi-associated nuclease 1 [Notothenia coriiceps] 1e-149; | ||||||
| NvERTx.4.126599 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | Q69ZT1 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|999979851|gb|KXJ19025.1|Fanconi-associated nuclease 1 [Exaiptasia pallida], 1e-78 |
| other nr_hits | gi|1005453634|ref|XP_015763977.1|PREDICTED: fanconi-associated nuclease 1-like [Acropora digitifera] 1e-71; gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera] 5e-70; gi|327284500|ref|XP_003226975.1|PREDICTED: fanconi-associated nuclease 1 [Anolis carolinensis] 4e-50; gi|705694806|ref|XP_010123642.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1, partial [Chlamydotis macqueenii] 8e-50; gi|543740761|ref|XP_005511262.1|PREDICTED: fanconi-associated nuclease 1 [Columba livia] 1e-49; gi|532029828|ref|XP_005357863.1|PREDICTED: fanconi-associated nuclease 1 [Microtus ochrogaster] 1e-49; gi|449270580|gb|EMC81239.1|Coiled-coil domain-containing protein MTMR15, partial [Columba livia] 1e-49; gi|556950934|ref|XP_005987805.1|PREDICTED: fanconi-associated nuclease 1 [Latimeria chalumnae] 2e-49; gi|699638631|ref|XP_009909560.1|PREDICTED: fanconi-associated nuclease 1 [Picoides pubescens] 2e-49; | ||||||
| NvERTx.4.126600 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | Q1LWH4 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera], 1e-59 |
| other nr_hits | gi|999979843|gb|KXJ19017.1|Fanconi-associated nuclease 1 [Exaiptasia pallida] 6e-47; gi|291232927|ref|XP_002736405.1|PREDICTED: fanconi-associated nuclease 1-like [Saccoglossus kowalevskii] 6e-42; gi|1126188815|ref|XP_019627245.1|PREDICTED: fanconi-associated nuclease 1-like [Branchiostoma belcheri] 2e-41; gi|1025381900|ref|XP_016400729.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus rhinocerous] 3e-39; gi|1101627130|ref|XP_018977325.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Cyprinus carpio] 1e-38; gi|1101627132|ref|XP_018977326.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Cyprinus carpio] 1e-38; gi|1025003863|ref|XP_016320959.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus anshuiensis] 1e-38; gi|1025003865|ref|XP_016320960.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Sinocyclocheilus anshuiensis] 1e-38; gi|160409999|sp|FAN1_DANRE|Q1LWH4.2|RecName: Full=Fanconi-associated nuclease 1; AltName: Full=FANCD2/FANCI-associated nuclease 1; AltName: Full=Myotubularin-related protein 15 2e-38; | ||||||
| NvERTx.4.126601 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | D2HNY3 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|999979851|gb|KXJ19025.1|Fanconi-associated nuclease 1 [Exaiptasia pallida], 2e-14 |
| other nr_hits | gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera] 9e-11; gi|1005453634|ref|XP_015763977.1|PREDICTED: fanconi-associated nuclease 1-like [Acropora digitifera] 1e-10; gi|1126188815|ref|XP_019627245.1|PREDICTED: fanconi-associated nuclease 1-like [Branchiostoma belcheri] 2e-05; | ||||||
| NvERTx.4.126602 | R-7 | 0.999959845635 | E-5 | 0.999999999957 | Q1LWH4 | Fanconi-associated nuclease 1 {ECO:0000250|UniProtKB:Q9Y2M0} | gi|1005453640|ref|XP_015763980.1|PREDICTED: LOW QUALITY PROTEIN: fanconi-associated nuclease 1-like [Acropora digitifera], 1e-59 |
| other nr_hits | gi|999979843|gb|KXJ19017.1|Fanconi-associated nuclease 1 [Exaiptasia pallida] 6e-47; gi|291232927|ref|XP_002736405.1|PREDICTED: fanconi-associated nuclease 1-like [Saccoglossus kowalevskii] 6e-42; gi|1126188815|ref|XP_019627245.1|PREDICTED: fanconi-associated nuclease 1-like [Branchiostoma belcheri] 2e-41; gi|1025381900|ref|XP_016400729.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus rhinocerous] 3e-39; gi|1101627130|ref|XP_018977325.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Cyprinus carpio] 1e-38; gi|1101627132|ref|XP_018977326.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Cyprinus carpio] 1e-38; gi|1025003863|ref|XP_016320959.1|PREDICTED: fanconi-associated nuclease 1-like isoform X1 [Sinocyclocheilus anshuiensis] 1e-38; gi|1025003865|ref|XP_016320960.1|PREDICTED: fanconi-associated nuclease 1-like isoform X2 [Sinocyclocheilus anshuiensis] 1e-38; gi|160409999|sp|FAN1_DANRE|Q1LWH4.2|RecName: Full=Fanconi-associated nuclease 1; AltName: Full=FANCD2/FANCI-associated nuclease 1; AltName: Full=Myotubularin-related protein 15 2e-38; | ||||||
| NvERTx.4.168360 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii], 2e-241 |
| other nr_hits | gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 3e-239; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 1e-238; gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri] 2e-238; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 7e-238; gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 2e-236; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 3e-236; gi|908479472|ref|XP_013091457.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Biomphalaria glabrata] 3e-233; gi|405953254|gb|EKC20952.1|Kinesin-like protein KIF27 [Crassostrea gigas] 3e-226; gi|919057531|ref|XP_013410613.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Lingula anatina] 3e-225; | ||||||
| NvERTx.4.168361 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii], 7e-246 |
| other nr_hits | gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 1e-243; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 4e-243; gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri] 9e-243; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 3e-242; gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 8e-241; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 1e-240; gi|908479472|ref|XP_013091457.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Biomphalaria glabrata] 1e-237; gi|405953254|gb|EKC20952.1|Kinesin-like protein KIF27 [Crassostrea gigas] 1e-230; gi|919057531|ref|XP_013410613.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Lingula anatina] 4e-229; | ||||||
| NvERTx.4.168362 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri], 7e-197 |
| other nr_hits | gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 6e-194; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 6e-194; gi|871236090|ref|XP_012938600.1|PREDICTED: kinesin-like protein KIF27 [Aplysia californica] 6e-194; gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii] 2e-193; gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 2e-193; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 9e-193; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 6e-192; gi|390337207|ref|XP_001194708.2|PREDICTED: kinesin-like protein KIF27 [Strongylocentrotus purpuratus] 2e-191; gi|919057531|ref|XP_013410613.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Lingula anatina] 5e-191; | ||||||
| NvERTx.4.168363 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri], 4e-226 |
| other nr_hits | gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 4e-224; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 1e-223; gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii] 4e-223; gi|919057531|ref|XP_013410613.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Lingula anatina] 7e-223; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 9e-223; gi|871236090|ref|XP_012938600.1|PREDICTED: kinesin-like protein KIF27 [Aplysia californica] 1e-221; gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 1e-220; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 1e-220; gi|390337207|ref|XP_001194708.2|PREDICTED: kinesin-like protein KIF27 [Strongylocentrotus purpuratus] 1e-219; | ||||||
| NvERTx.4.168364 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri], 1e-230 |
| other nr_hits | gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 1e-228; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 5e-228; gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii] 2e-227; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 3e-227; gi|919057531|ref|XP_013410613.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Lingula anatina] 1e-226; gi|871236090|ref|XP_012938600.1|PREDICTED: kinesin-like protein KIF27 [Aplysia californica] 5e-226; gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 5e-225; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 5e-225; gi|390337207|ref|XP_001194708.2|PREDICTED: kinesin-like protein KIF27 [Strongylocentrotus purpuratus] 5e-224; | ||||||
| NvERTx.4.168365 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri], 3e-201 |
| other nr_hits | gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 2e-198; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 2e-198; gi|871236090|ref|XP_012938600.1|PREDICTED: kinesin-like protein KIF27 [Aplysia californica] 2e-198; gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii] 7e-198; gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 9e-198; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 3e-197; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 2e-196; gi|390337207|ref|XP_001194708.2|PREDICTED: kinesin-like protein KIF27 [Strongylocentrotus purpuratus] 6e-196; gi|908479472|ref|XP_013091457.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Biomphalaria glabrata] 3e-195; | ||||||
| NvERTx.4.168366 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii], 2e-275 |
| other nr_hits | gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 2e-274; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 6e-274; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 4e-273; gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri] 5e-272; gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 2e-267; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 3e-267; gi|908479472|ref|XP_013091457.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Biomphalaria glabrata] 3e-264; gi|919057531|ref|XP_013410613.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Lingula anatina] 6e-261; gi|919057533|ref|XP_013410614.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Lingula anatina] 3e-256; | ||||||
| NvERTx.4.168367 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.168368 | R-1 | 0.47518231549 | E-5 | 0.999999999956 | Q86VH2 | Kinesin-like protein KIF27 | gi|585716683|ref|XP_006825564.1|PREDICTED: kinesin-like protein KIF27-like [Saccoglossus kowalevskii], 4e-271 |
| other nr_hits | gi|762141998|ref|XP_011454205.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Crassostrea gigas] 4e-270; gi|762142000|ref|XP_011454206.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Crassostrea gigas] 2e-269; gi|762141996|ref|XP_011454204.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Crassostrea gigas] 1e-268; gi|1126167055|ref|XP_019615374.1|PREDICTED: kinesin-like protein KIF27 [Branchiostoma belcheri] 1e-267; gi|908479467|ref|XP_013091455.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Biomphalaria glabrata] 5e-263; gi|908479469|ref|XP_013091456.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Biomphalaria glabrata] 7e-263; gi|908479472|ref|XP_013091457.1|PREDICTED: kinesin-like protein KIF27 isoform X3 [Biomphalaria glabrata] 7e-260; gi|919057531|ref|XP_013410613.1|PREDICTED: kinesin-like protein KIF27 isoform X1 [Lingula anatina] 4e-257; gi|919057533|ref|XP_013410614.1|PREDICTED: kinesin-like protein KIF27 isoform X2 [Lingula anatina] 2e-252; | ||||||
| NvERTx.4.40468 | R-7 | 0.999051950788 | E-5 | 0.999999999956 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.70136 | R-7 | 0.999051950788 | E-5 | 0.999999999956 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.70137 | R-7 | 0.999051950788 | E-5 | 0.999999999956 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.70138 | R-7 | 0.999051950788 | E-5 | 0.999999999956 | Q24297 | Small nuclear ribonucleoprotein F | gi|156392353|ref|XP_001636013.1|predicted protein [Nematostella vectensis], 9e-37 |
| other nr_hits | gi|1059398416|ref|XP_017772143.1|PREDICTED: small nuclear ribonucleoprotein F [Nicrophorus vespilloides] 2e-33; gi|291244974|ref|XP_002742366.1|PREDICTED: small nuclear ribonucleoprotein F-like [Saccoglossus kowalevskii] 3e-33; gi|919022875|ref|XP_013395334.1|PREDICTED: small nuclear ribonucleoprotein F [Lingula anatina] 5e-33; gi|961143115|ref|XP_014790194.1|PREDICTED: small nuclear ribonucleoprotein F [Octopus bimaculoides] 8e-33; gi|723219527|gb|AIX87794.1|cellular protein AbCp-93 [Androctonus bicolor] 1e-32; gi|908407719|ref|XP_013062243.1|PREDICTED: small nuclear ribonucleoprotein F [Biomphalaria glabrata] 1e-32; gi|957820135|ref|XP_014678163.1|PREDICTED: small nuclear ribonucleoprotein F, partial [Priapulus caudatus] 2e-32; gi|723219529|gb|AIX87795.1|cellular protein AbCp-94 [Androctonus bicolor] 3e-32; gi|260800049|ref|XP_002594949.1|hypothetical protein BRAFLDRAFT_271061 [Branchiostoma floridae] 4e-32; | ||||||
| NvERTx.4.70139 | R-7 | 0.999051950788 | E-5 | 0.999999999956 | Q24297 | Small nuclear ribonucleoprotein F | gi|156392353|ref|XP_001636013.1|predicted protein [Nematostella vectensis], 9e-37 |
| other nr_hits | gi|1059398416|ref|XP_017772143.1|PREDICTED: small nuclear ribonucleoprotein F [Nicrophorus vespilloides] 2e-33; gi|291244974|ref|XP_002742366.1|PREDICTED: small nuclear ribonucleoprotein F-like [Saccoglossus kowalevskii] 3e-33; gi|919022875|ref|XP_013395334.1|PREDICTED: small nuclear ribonucleoprotein F [Lingula anatina] 5e-33; gi|961143115|ref|XP_014790194.1|PREDICTED: small nuclear ribonucleoprotein F [Octopus bimaculoides] 8e-33; gi|723219527|gb|AIX87794.1|cellular protein AbCp-93 [Androctonus bicolor] 1e-32; gi|908407719|ref|XP_013062243.1|PREDICTED: small nuclear ribonucleoprotein F [Biomphalaria glabrata] 1e-32; gi|957820135|ref|XP_014678163.1|PREDICTED: small nuclear ribonucleoprotein F, partial [Priapulus caudatus] 2e-32; gi|723219529|gb|AIX87795.1|cellular protein AbCp-94 [Androctonus bicolor] 3e-32; gi|260800049|ref|XP_002594949.1|hypothetical protein BRAFLDRAFT_271061 [Branchiostoma floridae] 4e-32; | ||||||
| NvERTx.4.231824 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.132695 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | No_Uniprotmatch | No_Description | gi|156387546|ref|XP_001634264.1|predicted protein [Nematostella vectensis], 5e-14 |
| other nr_hits | NA | ||||||
| NvERTx.4.132697 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | No_Uniprotmatch | No_Description | gi|156387546|ref|XP_001634264.1|predicted protein [Nematostella vectensis], 6e-126 |
| other nr_hits | gi|1005426491|ref|XP_015751061.1|PREDICTED: trichohyalin-like [Acropora digitifera] 2e-18; | ||||||
| NvERTx.4.132698 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | E2C7F7_HARSA | Protein CASC3 {ECO:0000313|EMBL:EFN76144.1} | gi|156387546|ref|XP_001634264.1|predicted protein [Nematostella vectensis], 2e-196 |
| other nr_hits | gi|999986337|gb|KXJ25476.1|Protein CASC3 [Exaiptasia pallida] 1e-15; gi|1005426491|ref|XP_015751061.1|PREDICTED: trichohyalin-like [Acropora digitifera] 4e-11; gi|936599353|ref|XP_014213251.1|PREDICTED: bromodomain-containing protein 4-like [Copidosoma floridanum] 2e-09; gi|828199221|ref|XP_012554530.1|PREDICTED: probable serine/threonine-protein kinase DDB_G0280133 [Hydra vulgaris] 6e-09; gi|951575877|ref|XP_014485899.1|PREDICTED: protein CASC3 [Dinoponera quadriceps] 1e-08; gi|749797173|ref|XP_011152031.1|PREDICTED: protein CASC3 [Harpegnathos saltator] 1e-08; gi|665815609|ref|XP_008556469.1|PREDICTED: protein CASC3 [Microplitis demolitor] 2e-08; gi|795039433|ref|XP_011866275.1|PREDICTED: protein CASC3 isoform X1 [Vollenhovia emeryi] 3e-08; gi|795039436|ref|XP_011866276.1|PREDICTED: protein CASC3 isoform X2 [Vollenhovia emeryi] 3e-08; | ||||||
| NvERTx.4.132699 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | No_Uniprotmatch | No_Description | gi|156387546|ref|XP_001634264.1|predicted protein [Nematostella vectensis], 5e-36 |
| other nr_hits | gi|1005426491|ref|XP_015751061.1|PREDICTED: trichohyalin-like [Acropora digitifera] 4e-09; | ||||||
| NvERTx.4.132700 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | No_Uniprotmatch | No_Description | gi|156387546|ref|XP_001634264.1|predicted protein [Nematostella vectensis], 6e-126 |
| other nr_hits | gi|1005426491|ref|XP_015751061.1|PREDICTED: trichohyalin-like [Acropora digitifera] 8e-19; | ||||||
| NvERTx.4.132701 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | E2C7F7_HARSA | Protein CASC3 {ECO:0000313|EMBL:EFN76144.1} | gi|156387546|ref|XP_001634264.1|predicted protein [Nematostella vectensis], 1e-244 |
| other nr_hits | gi|1005426491|ref|XP_015751061.1|PREDICTED: trichohyalin-like [Acropora digitifera] 1e-18; gi|999986337|gb|KXJ25476.1|Protein CASC3 [Exaiptasia pallida] 2e-15; gi|936599353|ref|XP_014213251.1|PREDICTED: bromodomain-containing protein 4-like [Copidosoma floridanum] 2e-09; gi|828199221|ref|XP_012554530.1|PREDICTED: probable serine/threonine-protein kinase DDB_G0280133 [Hydra vulgaris] 8e-09; gi|951575877|ref|XP_014485899.1|PREDICTED: protein CASC3 [Dinoponera quadriceps] 2e-08; gi|749797173|ref|XP_011152031.1|PREDICTED: protein CASC3 [Harpegnathos saltator] 2e-08; gi|665815609|ref|XP_008556469.1|PREDICTED: protein CASC3 [Microplitis demolitor] 3e-08; gi|795039433|ref|XP_011866275.1|PREDICTED: protein CASC3 isoform X1 [Vollenhovia emeryi] 4e-08; gi|795039436|ref|XP_011866276.1|PREDICTED: protein CASC3 isoform X2 [Vollenhovia emeryi] 4e-08; | ||||||
| NvERTx.4.132702 | R-7 | 0.999736960978 | E-5 | 0.999999999955 | E2C7F7_HARSA | Protein CASC3 {ECO:0000313|EMBL:EFN76144.1} | gi|156387546|ref|XP_001634264.1|predicted protein [Nematostella vectensis], 1e-244 |
| other nr_hits | gi|1005426491|ref|XP_015751061.1|PREDICTED: trichohyalin-like [Acropora digitifera] 4e-18; gi|999986337|gb|KXJ25476.1|Protein CASC3 [Exaiptasia pallida] 2e-15; gi|936599353|ref|XP_014213251.1|PREDICTED: bromodomain-containing protein 4-like [Copidosoma floridanum] 2e-09; gi|828199221|ref|XP_012554530.1|PREDICTED: probable serine/threonine-protein kinase DDB_G0280133 [Hydra vulgaris] 8e-09; gi|951575877|ref|XP_014485899.1|PREDICTED: protein CASC3 [Dinoponera quadriceps] 2e-08; gi|749797173|ref|XP_011152031.1|PREDICTED: protein CASC3 [Harpegnathos saltator] 2e-08; gi|665815609|ref|XP_008556469.1|PREDICTED: protein CASC3 [Microplitis demolitor] 3e-08; gi|795039433|ref|XP_011866275.1|PREDICTED: protein CASC3 isoform X1 [Vollenhovia emeryi] 4e-08; gi|795039436|ref|XP_011866276.1|PREDICTED: protein CASC3 isoform X2 [Vollenhovia emeryi] 4e-08; | ||||||
| NvERTx.4.154447 | R-7 | 0.898569320253 | E-5 | 0.999999999955 | Q91VH6 | Protein MEMO1 | gi|156368571|ref|XP_001627766.1|predicted protein [Nematostella vectensis], 2e-171 |
| other nr_hits | gi|999978310|gb|KXJ17484.1|Protein MEMO1 [Exaiptasia pallida] 5e-124; gi|1005443122|ref|XP_015758905.1|PREDICTED: protein MEMO1-like [Acropora digitifera] 1e-114; gi|1036830083|ref|XP_016976615.1|PREDICTED: protein MEMO1 [Drosophila rhopaloa] 1e-111; gi|195446372|ref|XP_002070750.1|uncharacterized protein Dwil_GK10855 [Drosophila willistoni] 4e-111; gi|1036058269|ref|XP_016932235.1|PREDICTED: protein MEMO1-like [Drosophila suzukii] 1e-110; gi|1037098463|ref|XP_017084434.1|PREDICTED: protein MEMO1 [Drosophila eugracilis] 1e-110; gi|1036719535|ref|XP_017134412.1|PREDICTED: protein MEMO1 [Drosophila elegans] 1e-110; gi|194743160|ref|XP_001954068.1|uncharacterized protein Dana_GF18089 [Drosophila ananassae] 1e-110; gi|1036753547|ref|XP_017039943.1|PREDICTED: protein MEMO1 [Drosophila ficusphila] 2e-110; | ||||||
| NvERTx.4.154456 | R-7 | 0.898569320253 | E-5 | 0.999999999955 | Q91VH6 | Protein MEMO1 | gi|156368571|ref|XP_001627766.1|predicted protein [Nematostella vectensis], 2e-171 |
| other nr_hits | gi|999978310|gb|KXJ17484.1|Protein MEMO1 [Exaiptasia pallida] 5e-124; gi|1005443122|ref|XP_015758905.1|PREDICTED: protein MEMO1-like [Acropora digitifera] 1e-114; gi|1036830083|ref|XP_016976615.1|PREDICTED: protein MEMO1 [Drosophila rhopaloa] 1e-111; gi|195446372|ref|XP_002070750.1|uncharacterized protein Dwil_GK10855 [Drosophila willistoni] 4e-111; gi|1036058269|ref|XP_016932235.1|PREDICTED: protein MEMO1-like [Drosophila suzukii] 1e-110; gi|1037098463|ref|XP_017084434.1|PREDICTED: protein MEMO1 [Drosophila eugracilis] 1e-110; gi|1036719535|ref|XP_017134412.1|PREDICTED: protein MEMO1 [Drosophila elegans] 1e-110; gi|194743160|ref|XP_001954068.1|uncharacterized protein Dana_GF18089 [Drosophila ananassae] 1e-110; gi|1036753547|ref|XP_017039943.1|PREDICTED: protein MEMO1 [Drosophila ficusphila] 2e-110; | ||||||
| NvERTx.4.154457 | R-7 | 0.898569320253 | E-5 | 0.999999999955 | Q4R6D9 | Protein MEMO1 | gi|156368571|ref|XP_001627766.1|predicted protein [Nematostella vectensis], 2e-66 |
| other nr_hits | gi|999978310|gb|KXJ17484.1|Protein MEMO1 [Exaiptasia pallida] 2e-47; gi|1005443122|ref|XP_015758905.1|PREDICTED: protein MEMO1-like [Acropora digitifera] 3e-41; gi|390332252|ref|XP_791245.2|PREDICTED: protein MEMO1 [Strongylocentrotus purpuratus] 2e-38; gi|1037098463|ref|XP_017084434.1|PREDICTED: protein MEMO1 [Drosophila eugracilis] 4e-38; gi|1036830083|ref|XP_016976615.1|PREDICTED: protein MEMO1 [Drosophila rhopaloa] 9e-38; gi|1036719535|ref|XP_017134412.1|PREDICTED: protein MEMO1 [Drosophila elegans] 1e-37; gi|195997049|ref|XP_002108393.1|hypothetical protein TRIADDRAFT_37128 [Trichoplax adhaerens] 1e-37; gi|676596378|gb|KFO82209.1|Protein MEMO1, partial [Cuculus canorus] 2e-37; gi|195571271|ref|XP_002103627.1|uncharacterized protein Dsimw501_GD20526 [Drosophila simulans] 2e-37; | ||||||
| NvERTx.4.154458 | R-7 | 0.898569320253 | E-5 | 0.999999999955 | Q91VH6 | Protein MEMO1 | gi|156368571|ref|XP_001627766.1|predicted protein [Nematostella vectensis], 2e-171 |
| other nr_hits | gi|999978310|gb|KXJ17484.1|Protein MEMO1 [Exaiptasia pallida] 5e-124; gi|1005443122|ref|XP_015758905.1|PREDICTED: protein MEMO1-like [Acropora digitifera] 1e-114; gi|1036830083|ref|XP_016976615.1|PREDICTED: protein MEMO1 [Drosophila rhopaloa] 1e-111; gi|195446372|ref|XP_002070750.1|uncharacterized protein Dwil_GK10855 [Drosophila willistoni] 4e-111; gi|1036058269|ref|XP_016932235.1|PREDICTED: protein MEMO1-like [Drosophila suzukii] 1e-110; gi|1037098463|ref|XP_017084434.1|PREDICTED: protein MEMO1 [Drosophila eugracilis] 1e-110; gi|1036719535|ref|XP_017134412.1|PREDICTED: protein MEMO1 [Drosophila elegans] 1e-110; gi|194743160|ref|XP_001954068.1|uncharacterized protein Dana_GF18089 [Drosophila ananassae] 1e-110; gi|1036753547|ref|XP_017039943.1|PREDICTED: protein MEMO1 [Drosophila ficusphila] 2e-110; | ||||||
| NvERTx.4.154459 | R-7 | 0.898569320253 | E-5 | 0.999999999955 | Q91VH6 | Protein MEMO1 | gi|156368571|ref|XP_001627766.1|predicted protein [Nematostella vectensis], 2e-171 |
| other nr_hits | gi|999978310|gb|KXJ17484.1|Protein MEMO1 [Exaiptasia pallida] 5e-124; gi|1005443122|ref|XP_015758905.1|PREDICTED: protein MEMO1-like [Acropora digitifera] 1e-114; gi|1036830083|ref|XP_016976615.1|PREDICTED: protein MEMO1 [Drosophila rhopaloa] 1e-111; gi|195446372|ref|XP_002070750.1|uncharacterized protein Dwil_GK10855 [Drosophila willistoni] 4e-111; gi|1036058269|ref|XP_016932235.1|PREDICTED: protein MEMO1-like [Drosophila suzukii] 1e-110; gi|1037098463|ref|XP_017084434.1|PREDICTED: protein MEMO1 [Drosophila eugracilis] 1e-110; gi|1036719535|ref|XP_017134412.1|PREDICTED: protein MEMO1 [Drosophila elegans] 1e-110; gi|194743160|ref|XP_001954068.1|uncharacterized protein Dana_GF18089 [Drosophila ananassae] 1e-110; gi|1036753547|ref|XP_017039943.1|PREDICTED: protein MEMO1 [Drosophila ficusphila] 2e-110; | ||||||
