Explore Mfuzz Clusters
Mfuzz cluster
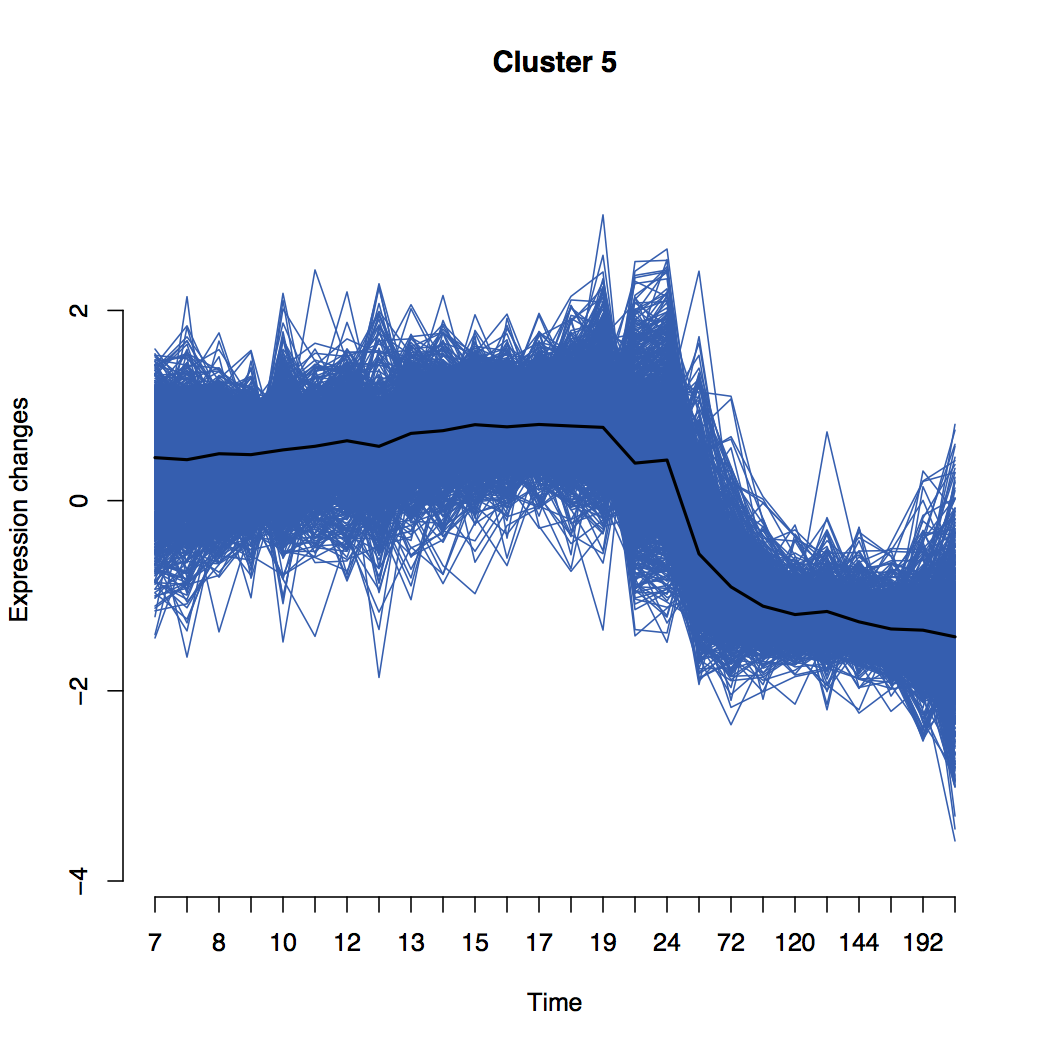
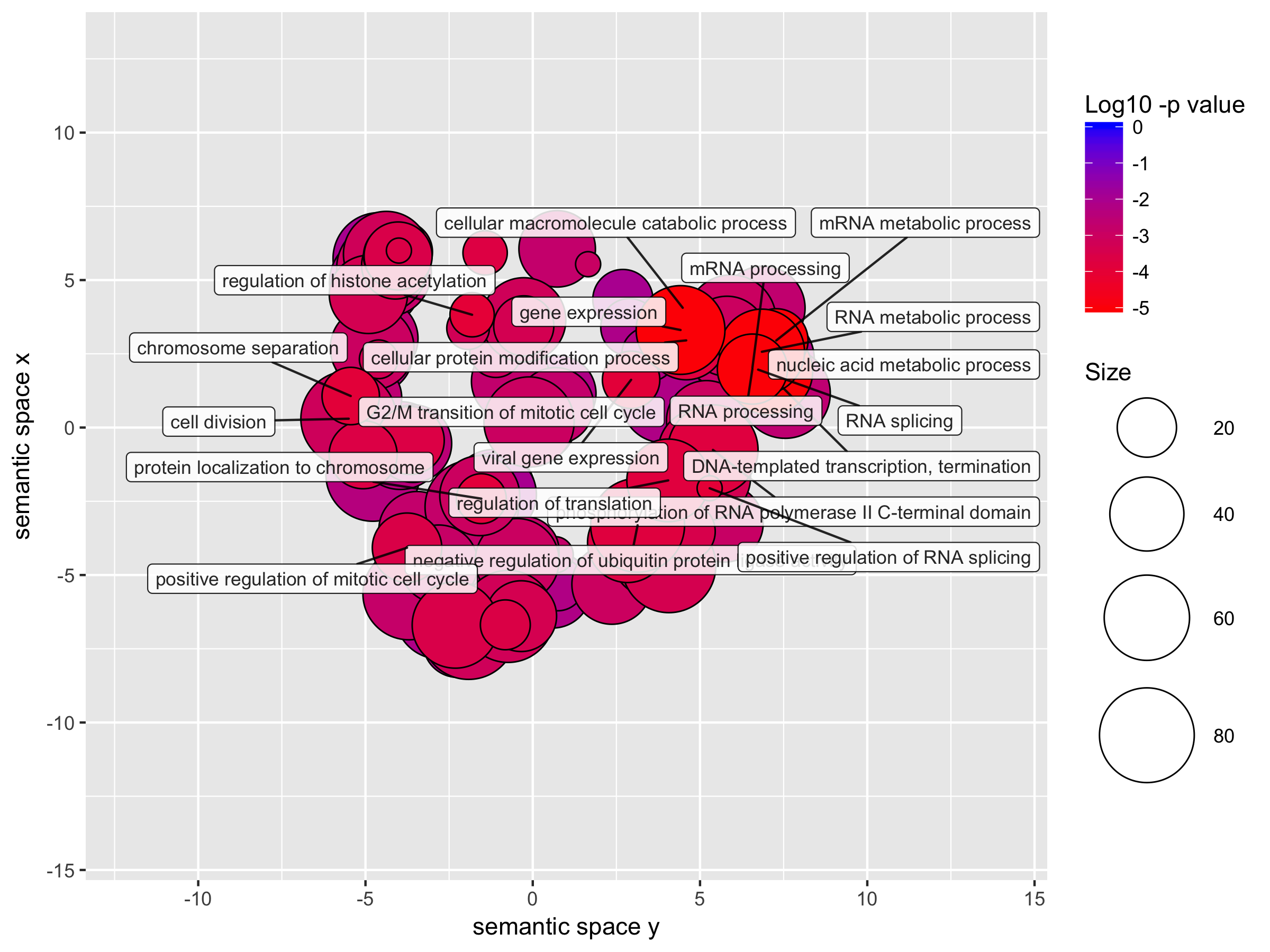
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.118343 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | Q9U5M4 | Tropomyosin-2 | gi|156389265|ref|XP_001634912.1|predicted protein [Nematostella vectensis], 1e-90 |
| other nr_hits | gi|95102190|gb|ABF51018.1|tropomyosin 2 [Nematostella vectensis] 5e-90; gi|156389267|ref|XP_001634913.1|predicted protein [Nematostella vectensis] 1e-35; gi|999970609|gb|KXJ09783.1|Tropomyosin-2 [Exaiptasia pallida] 1e-30; gi|1005431290|ref|XP_015753349.1|PREDICTED: tropomyosin-like [Acropora digitifera] 1e-23; gi|999978819|gb|KXJ17993.1|Tropomyosin [Exaiptasia pallida] 2e-17; gi|156379976|ref|XP_001631731.1|predicted protein [Nematostella vectensis] 1e-16; gi|221132153|ref|XP_002165211.1|PREDICTED: tropomyosin-1-like [Hydra vulgaris] 1e-16; gi|42559896|sp|TPM2_PODCA|Q9U5M4.1|RecName: Full=Tropomyosin-2 >CAB55601.1 tropomyosin [Podocoryna carnea] 3e-16; gi|321478982|gb|EFX89938.1|hypothetical protein DAPPUDRAFT_230081 [Daphnia pulex] 5e-16; | ||||||
| NvERTx.4.118344 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.118345 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.118348 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | gi|999983256|gb|KXJ22397.1|hypothetical protein AC249_AIPGENE27418 [Exaiptasia pallida], 6e-23 |
| other nr_hits | gi|1005477967|ref|XP_015775739.1|PREDICTED: uncharacterized protein LOC107353864 [Acropora digitifera] 3e-18; gi|999983294|gb|KXJ22435.1|hypothetical protein AC249_AIPGENE27386 [Exaiptasia pallida] 5e-17; gi|156401169|ref|XP_001639164.1|predicted protein [Nematostella vectensis] 7e-16; | ||||||
| NvERTx.4.118354 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | gi|999983256|gb|KXJ22397.1|hypothetical protein AC249_AIPGENE27418 [Exaiptasia pallida], 1e-34 |
| other nr_hits | gi|999983294|gb|KXJ22435.1|hypothetical protein AC249_AIPGENE27386 [Exaiptasia pallida] 1e-16; gi|156401169|ref|XP_001639164.1|predicted protein [Nematostella vectensis] 4e-12; | ||||||
| NvERTx.4.164714 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.164715 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.164717 | R-2 | 0.420605605623 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.55421 | R-5 | 0.98716419314 | E-5 | 0.999999999997 | Q9R0Q9 | Mannose-P-dolichol utilization defect 1 protein | gi|156395756|ref|XP_001637276.1|predicted protein [Nematostella vectensis], 2e-130 |
| other nr_hits | gi|1005458683|ref|XP_015766477.1|PREDICTED: mannose-P-dolichol utilization defect 1 protein-like isoform X1 [Acropora digitifera] 8e-77; gi|999970104|gb|KXJ09278.1|Mannose-P-dolichol utilization defect 1 protein [Exaiptasia pallida] 7e-73; gi|585656894|ref|XP_002734165.2|PREDICTED: mannose-P-dolichol utilization defect 1 protein-like [Saccoglossus kowalevskii] 3e-64; gi|505787104|ref|XP_004604919.1|PREDICTED: mannose-P-dolichol utilization defect 1 protein [Sorex araneus] 5e-61; gi|641723760|ref|XP_008151732.1|PREDICTED: mannose-P-dolichol utilization defect 1 protein [Eptesicus fuscus] 8e-61; gi|524926731|ref|XP_005067600.1|PREDICTED: mannose-P-dolichol utilization defect 1 protein isoform X1 [Mesocricetus auratus] 1e-60; gi|554569106|ref|XP_005878816.1|PREDICTED: mannose-P-dolichol utilization defect 1 protein [Myotis brandtii] 3e-60; gi|558201390|ref|XP_006105573.1|PREDICTED: mannose-P-dolichol utilization defect 1 protein [Myotis lucifugus] 4e-60; gi|507953795|ref|XP_004684702.1|PREDICTED: mannose-P-dolichol utilization defect 1 protein isoform X1 [Condylura cristata] 4e-60; | ||||||
| NvERTx.4.84731 | R-5 | 0.98716419314 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.18630 | R-7 | 0.999982908553 | E-5 | 0.999999999997 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.123457 | R-7 | 0.999982908553 | E-5 | 0.999999999997 | Q3SZZ0 | Ribosome biogenesis protein BRX1 homolog | gi|156392325|ref|XP_001635999.1|predicted protein [Nematostella vectensis], 1e-142 |
| other nr_hits | gi|957823431|ref|XP_014661421.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Priapulus caudatus] 3e-99; gi|524885374|ref|XP_005099295.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Aplysia californica] 5e-99; gi|919025760|ref|XP_013396650.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Lingula anatina] 2e-98; gi|830014072|ref|XP_012613584.1|ribosome biogenesis protein BRX1 homolog isoform X1 [Microcebus murinus] 1e-97; gi|908441198|ref|XP_013079143.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Biomphalaria glabrata] 3e-97; gi|826282969|ref|XP_012495988.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Propithecus coquereli] 6e-97; gi|635027677|ref|XP_007959524.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Chlorocebus sabaeus] 6e-97; gi|987941882|ref|XP_006761828.2|PREDICTED: ribosome biogenesis protein BRX1 homolog isoform X1 [Myotis davidii] 1e-96; gi|395840263|ref|XP_003792982.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Otolemur garnettii] 1e-96; | ||||||
| NvERTx.4.209158 | R-7 | 0.999982908553 | E-5 | 0.999999999997 | Q3SZZ0 | Ribosome biogenesis protein BRX1 homolog | gi|156392325|ref|XP_001635999.1|predicted protein [Nematostella vectensis], 1e-31 |
| other nr_hits | gi|919025760|ref|XP_013396650.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Lingula anatina] 7e-23; gi|291395226|ref|XP_002714177.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Oryctolagus cuniculus] 1e-22; gi|970706468|ref|XP_015090947.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Vicugna pacos] 2e-22; gi|946689071|ref|XP_014423323.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Camelus ferus] 2e-22; gi|744557478|ref|XP_010975911.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Camelus dromedarius] 2e-22; gi|743720119|ref|XP_010953845.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Camelus bactrianus] 2e-22; gi|528753495|gb|EPY73154.1|ribosome biogenesis protein BRX1-like protein [Camelus ferus] 2e-22; gi|902895734|ref|XP_013037169.1|PREDICTED: ribosome biogenesis protein BRX1 homolog [Anser cygnoides domesticus] 2e-22; gi|874466569|ref|XP_005031951.2|PREDICTED: LOW QUALITY PROTEIN: ribosome biogenesis protein BRX1 homolog, partial [Anas platyrhynchos] 2e-22; | ||||||
| NvERTx.4.84175 | R-5 | 0.997701024526 | E-5 | 0.999999999996 | Q6P926 | Spermatogenesis-associated protein 24 | gi|156402736|ref|XP_001639746.1|predicted protein [Nematostella vectensis], 1e-45 |
| other nr_hits | gi|999975471|gb|KXJ14645.1|Spermatogenesis-associated protein 24 [Exaiptasia pallida] 2e-27; gi|1005425106|ref|XP_015779355.1|PREDICTED: spermatogenesis-associated protein 24-like isoform X1 [Acropora digitifera] 3e-18; gi|1005425108|ref|XP_015779361.1|PREDICTED: spermatogenesis-associated protein 24-like isoform X2 [Acropora digitifera] 3e-18; gi|291228056|ref|XP_002733994.1|PREDICTED: spermatogenesis-associated protein 24-like [Saccoglossus kowalevskii] 2e-14; gi|1126204689|ref|XP_019635845.1|PREDICTED: LOW QUALITY PROTEIN: spermatogenesis-associated protein 24-like [Branchiostoma belcheri] 5e-12; gi|196009352|ref|XP_002114541.1|hypothetical protein TRIADDRAFT_58473 [Trichoplax adhaerens] 1e-11; gi|632957530|ref|XP_007894530.1|PREDICTED: spermatogenesis-associated protein 24 [Callorhinchus milii] 3e-10; gi|558103855|ref|XP_006110411.1|PREDICTED: spermatogenesis-associated protein 24 [Pelodiscus sinensis] 4e-10; gi|780130201|ref|XP_011679172.1|PREDICTED: spermatogenesis-associated protein 24 isoform X2 [Strongylocentrotus purpuratus] 6e-10; | ||||||
| NvERTx.4.84180 | R-5 | 0.997701024526 | E-5 | 0.999999999996 | Q6P926 | Spermatogenesis-associated protein 24 | gi|156402736|ref|XP_001639746.1|predicted protein [Nematostella vectensis], 1e-82 |
| other nr_hits | gi|999975471|gb|KXJ14645.1|Spermatogenesis-associated protein 24 [Exaiptasia pallida] 2e-59; gi|1005425106|ref|XP_015779355.1|PREDICTED: spermatogenesis-associated protein 24-like isoform X1 [Acropora digitifera] 6e-55; gi|1005425108|ref|XP_015779361.1|PREDICTED: spermatogenesis-associated protein 24-like isoform X2 [Acropora digitifera] 3e-44; gi|291228056|ref|XP_002733994.1|PREDICTED: spermatogenesis-associated protein 24-like [Saccoglossus kowalevskii] 1e-43; gi|632957530|ref|XP_007894530.1|PREDICTED: spermatogenesis-associated protein 24 [Callorhinchus milii] 3e-39; gi|1126204689|ref|XP_019635845.1|PREDICTED: LOW QUALITY PROTEIN: spermatogenesis-associated protein 24-like [Branchiostoma belcheri] 1e-37; gi|260786107|ref|XP_002588100.1|hypothetical protein BRAFLDRAFT_87619 [Branchiostoma floridae] 2e-37; gi|641768050|ref|XP_008168625.1|PREDICTED: spermatogenesis-associated protein 24 isoform X2 [Chrysemys picta bellii] 1e-35; gi|919052371|ref|XP_013408147.1|PREDICTED: spermatogenesis-associated protein 24-like [Lingula anatina] 2e-35; | ||||||
| NvERTx.4.221185 | R-5 | 0.997701024526 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | gi|156402736|ref|XP_001639746.1|predicted protein [Nematostella vectensis], 5e-05 |
| other nr_hits | NA | ||||||
| NvERTx.4.221929 | R-5 | 0.997701024526 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.133927 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | P79303 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 2e-30 |
| other nr_hits | gi|999983140|gb|KXJ22283.1|UTP--glucose-1-phosphate uridylyltransferase [Exaiptasia pallida] 6e-26; gi|1129971748|ref|XP_019743667.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X2 [Hippocampus comes] 3e-24; gi|1129971746|ref|XP_019743666.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Hippocampus comes] 3e-24; gi|1101626278|ref|XP_018976898.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Cyprinus carpio] 3e-24; gi|1101626280|ref|XP_018976899.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X2 [Cyprinus carpio] 3e-24; gi|765142691|ref|XP_011482485.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like isoform X1 [Oryzias latipes] 4e-24; gi|432902856|ref|XP_004077045.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like isoform X2 [Oryzias latipes] 4e-24; gi|1108924939|ref|XP_019116431.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Larimichthys crocea] 7e-24; gi|1083293254|ref|XP_018595549.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Scleropages formosus] 7e-24; | ||||||
| NvERTx.4.133928 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | Q07130 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 4e-287 |
| other nr_hits | gi|1073530231|gb|JAT82227.1|hypothetical protein g.13527 [Pectinophora gossypiella] 9e-184; gi|1073532925|gb|JAT83574.1|hypothetical protein g.13530 [Pectinophora gossypiella] 9e-184; gi|1073538593|gb|JAT86408.1|hypothetical protein g.13529 [Pectinophora gossypiella] 9e-184; gi|1073547613|gb|JAT90918.1|hypothetical protein g.13528 [Pectinophora gossypiella] 9e-184; gi|224496086|ref|NP_001139029.1|UDP-glucose pyrophosphorylase 2a [Danio rerio] 1e-182; gi|528469138|ref|XP_005155455.1|PREDICTED: UDP-glucose pyrophosphorylase 2a isoform X1 [Danio rerio] 1e-182; gi|357609270|gb|EHJ66377.1|hypothetical protein KGM_06440 [Danaus plexippus] 2e-182; gi|1024936856|ref|XP_016325590.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Sinocyclocheilus anshuiensis] 7e-182; gi|617463907|ref|XP_007572987.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Poecilia formosa] 2e-180; | ||||||
| NvERTx.4.133929 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | Q07130 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 1e-282 |
| other nr_hits | gi|1073530231|gb|JAT82227.1|hypothetical protein g.13527 [Pectinophora gossypiella] 9e-184; gi|1073532925|gb|JAT83574.1|hypothetical protein g.13530 [Pectinophora gossypiella] 9e-184; gi|1073538593|gb|JAT86408.1|hypothetical protein g.13529 [Pectinophora gossypiella] 9e-184; gi|1073547613|gb|JAT90918.1|hypothetical protein g.13528 [Pectinophora gossypiella] 9e-184; gi|224496086|ref|NP_001139029.1|UDP-glucose pyrophosphorylase 2a [Danio rerio] 1e-182; gi|528469138|ref|XP_005155455.1|PREDICTED: UDP-glucose pyrophosphorylase 2a isoform X1 [Danio rerio] 1e-182; gi|357609270|gb|EHJ66377.1|hypothetical protein KGM_06440 [Danaus plexippus] 2e-182; gi|1024936856|ref|XP_016325590.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Sinocyclocheilus anshuiensis] 6e-182; gi|585723852|ref|XP_006813715.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like isoform X1 [Saccoglossus kowalevskii] 2e-180; | ||||||
| NvERTx.4.133930 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | O35156 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 1e-47 |
| other nr_hits | gi|1073530231|gb|JAT82227.1|hypothetical protein g.13527 [Pectinophora gossypiella] 1e-35; gi|1073532925|gb|JAT83574.1|hypothetical protein g.13530 [Pectinophora gossypiella] 1e-35; gi|1073538593|gb|JAT86408.1|hypothetical protein g.13529 [Pectinophora gossypiella] 1e-35; gi|1073547613|gb|JAT90918.1|hypothetical protein g.13528 [Pectinophora gossypiella] 1e-35; gi|780077458|ref|XP_011672365.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Strongylocentrotus purpuratus] 1e-35; gi|943949819|ref|XP_014357929.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Papilio machaon] 1e-35; gi|943949824|ref|XP_014357931.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X3 [Papilio machaon] 1e-35; gi|943949826|ref|XP_014357932.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X4 [Papilio machaon] 1e-35; gi|943949821|ref|XP_014357930.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X2 [Papilio machaon] 1e-35; | ||||||
| NvERTx.4.133931 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | Q07130 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 4e-287 |
| other nr_hits | gi|1073530231|gb|JAT82227.1|hypothetical protein g.13527 [Pectinophora gossypiella] 9e-184; gi|1073532925|gb|JAT83574.1|hypothetical protein g.13530 [Pectinophora gossypiella] 9e-184; gi|1073538593|gb|JAT86408.1|hypothetical protein g.13529 [Pectinophora gossypiella] 9e-184; gi|1073547613|gb|JAT90918.1|hypothetical protein g.13528 [Pectinophora gossypiella] 9e-184; gi|224496086|ref|NP_001139029.1|UDP-glucose pyrophosphorylase 2a [Danio rerio] 1e-182; gi|528469138|ref|XP_005155455.1|PREDICTED: UDP-glucose pyrophosphorylase 2a isoform X1 [Danio rerio] 1e-182; gi|357609270|gb|EHJ66377.1|hypothetical protein KGM_06440 [Danaus plexippus] 2e-182; gi|1024936856|ref|XP_016325590.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Sinocyclocheilus anshuiensis] 7e-182; gi|617463907|ref|XP_007572987.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Poecilia formosa] 2e-180; | ||||||
| NvERTx.4.133932 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | O35156 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 1e-47 |
| other nr_hits | gi|1073530231|gb|JAT82227.1|hypothetical protein g.13527 [Pectinophora gossypiella] 1e-35; gi|1073532925|gb|JAT83574.1|hypothetical protein g.13530 [Pectinophora gossypiella] 1e-35; gi|1073538593|gb|JAT86408.1|hypothetical protein g.13529 [Pectinophora gossypiella] 1e-35; gi|1073547613|gb|JAT90918.1|hypothetical protein g.13528 [Pectinophora gossypiella] 1e-35; gi|780077458|ref|XP_011672365.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Strongylocentrotus purpuratus] 1e-35; gi|943949819|ref|XP_014357929.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Papilio machaon] 1e-35; gi|943949824|ref|XP_014357931.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X3 [Papilio machaon] 1e-35; gi|943949826|ref|XP_014357932.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X4 [Papilio machaon] 1e-35; gi|943949821|ref|XP_014357930.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X2 [Papilio machaon] 1e-35; | ||||||
| NvERTx.4.133933 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | Q07130 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 1e-282 |
| other nr_hits | gi|1073530231|gb|JAT82227.1|hypothetical protein g.13527 [Pectinophora gossypiella] 9e-184; gi|1073532925|gb|JAT83574.1|hypothetical protein g.13530 [Pectinophora gossypiella] 9e-184; gi|1073538593|gb|JAT86408.1|hypothetical protein g.13529 [Pectinophora gossypiella] 9e-184; gi|1073547613|gb|JAT90918.1|hypothetical protein g.13528 [Pectinophora gossypiella] 9e-184; gi|224496086|ref|NP_001139029.1|UDP-glucose pyrophosphorylase 2a [Danio rerio] 1e-182; gi|528469138|ref|XP_005155455.1|PREDICTED: UDP-glucose pyrophosphorylase 2a isoform X1 [Danio rerio] 1e-182; gi|357609270|gb|EHJ66377.1|hypothetical protein KGM_06440 [Danaus plexippus] 2e-182; gi|1024936856|ref|XP_016325590.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Sinocyclocheilus anshuiensis] 6e-182; gi|585723852|ref|XP_006813715.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like isoform X1 [Saccoglossus kowalevskii] 2e-180; | ||||||
| NvERTx.4.133934 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.207255 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.213689 | R-1 | 0.936310404458 | E-5 | 0.999999999996 | O35156 | UTP--glucose-1-phosphate uridylyltransferase | gi|156405246|ref|XP_001640643.1|predicted protein [Nematostella vectensis], 1e-09 |
| other nr_hits | gi|1025215163|ref|XP_016429849.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Sinocyclocheilus rhinocerous] 3e-09; gi|1025066346|ref|XP_016335980.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Sinocyclocheilus anshuiensis] 3e-09; gi|1020412171|ref|XP_016124801.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Sinocyclocheilus grahami] 3e-09; gi|926636695|ref|XP_013784292.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase-like [Limulus polyphemus] 3e-09; gi|1158923643|ref|XP_020309643.1|UTP--glucose-1-phosphate uridylyltransferase-like isoform X1 [Oncorhynchus kisutch] 3e-09; gi|1158923645|ref|XP_020309644.1|UTP--glucose-1-phosphate uridylyltransferase-like isoform X2 [Oncorhynchus kisutch] 3e-09; gi|1158851667|ref|XP_020336916.1|UTP--glucose-1-phosphate uridylyltransferase-like isoform X2 [Oncorhynchus kisutch] 3e-09; gi|1158851665|ref|XP_020336907.1|UTP--glucose-1-phosphate uridylyltransferase-like isoform X1 [Oncorhynchus kisutch] 3e-09; gi|1003756540|ref|XP_015712042.1|PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Coturnix japonica] 3e-09; | ||||||
| NvERTx.4.211025 | R-7 | 0.965764943132 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.225148 | R-7 | 0.965764943132 | E-5 | 0.999999999996 | Q5C9L6 | (S)-coclaurine N-methyltransferase | gi|156408083|ref|XP_001641686.1|predicted protein [Nematostella vectensis], 1e-194 |
| other nr_hits | gi|999987376|gb|KXJ26493.1|(S)-coclaurine N-methyltransferase [Exaiptasia pallida] 3e-149; gi|1005458113|ref|XP_015766181.1|PREDICTED: (S)-coclaurine N-methyltransferase-like [Acropora digitifera] 6e-124; gi|762086443|ref|XP_011425493.1|PREDICTED: (S)-coclaurine N-methyltransferase [Crassostrea gigas] 5e-123; gi|524877504|ref|XP_005095452.1|PREDICTED: (S)-coclaurine N-methyltransferase-like [Aplysia californica] 1e-122; gi|908406315|ref|XP_013092484.1|PREDICTED: (S)-coclaurine N-methyltransferase-like [Biomphalaria glabrata] 3e-122; gi|676434800|ref|XP_009047348.1|hypothetical protein LOTGIDRAFT_230578 [Lottia gigantea] 4e-122; gi|918998856|ref|XP_013397285.1|PREDICTED: (S)-coclaurine N-methyltransferase-like [Lingula anatina] 1e-120; gi|585712954|ref|XP_002741514.2|PREDICTED: (S)-coclaurine N-methyltransferase-like [Saccoglossus kowalevskii] 1e-118; gi|1126185107|ref|XP_019625206.1|PREDICTED: (S)-coclaurine N-methyltransferase-like [Branchiostoma belcheri] 2e-115; | ||||||
| NvERTx.4.70140 | R-7 | 0.999816596604 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.70141 | R-7 | 0.999816596604 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.70142 | R-7 | 0.999816596604 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | gi|156398849|ref|XP_001638400.1|predicted protein [Nematostella vectensis], 6e-112 |
| other nr_hits | gi|1005431671|ref|XP_015753511.1|PREDICTED: bcl-2-associated transcription factor 1-like [Acropora digitifera] 3e-12; | ||||||
| NvERTx.4.91817 | R-7 | 0.999669348676 | E-5 | 0.999999999996 | Q6NWD4 | Mitochondrial import inner membrane translocase subunit TIM50 | gi|999980690|gb|KXJ19864.1|Mitochondrial import inner membrane translocase subunit TIM50 [Exaiptasia pallida], 2e-126 |
| other nr_hits | gi|156368410|ref|XP_001627687.1|predicted protein [Nematostella vectensis] 1e-122; gi|828218050|ref|XP_012560847.1|PREDICTED: mitochondrial import inner membrane translocase subunit TIM50-like [Hydra vulgaris] 4e-81; gi|260834727|ref|XP_002612361.1|hypothetical protein BRAFLDRAFT_265448 [Branchiostoma floridae] 2e-77; gi|780087525|ref|XP_786728.4|PREDICTED: mitochondrial import inner membrane translocase subunit TIM50 [Strongylocentrotus purpuratus] 2e-76; gi|1126192299|ref|XP_019629142.1|PREDICTED: mitochondrial import inner membrane translocase subunit TIM50-like [Branchiostoma belcheri] 1e-75; gi|831302899|ref|XP_012683519.1|PREDICTED: mitochondrial import inner membrane translocase subunit TIM50 isoform X2 [Clupea harengus] 1e-75; gi|1101618461|ref|XP_018972918.1|PREDICTED: mitochondrial import inner membrane translocase subunit TIM50 [Cyprinus carpio] 3e-75; gi|1024973727|ref|XP_016308381.1|PREDICTED: mitochondrial import inner membrane translocase subunit TIM50 [Sinocyclocheilus anshuiensis] 4e-75; gi|1158916392|ref|XP_020363314.1|mitochondrial import inner membrane translocase subunit TIM50-like [Oncorhynchus kisutch] 5e-75; | ||||||
| NvERTx.4.15457 | R-1 | 0.870259999329 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.149810 | R-1 | 0.870259999329 | E-5 | 0.999999999996 | P17225 | Polypyrimidine tract-binding protein 1 | gi|156392443|ref|XP_001636058.1|predicted protein [Nematostella vectensis], 1e-124 |
| other nr_hits | gi|999984893|gb|KXJ24034.1|Polypyrimidine tract-binding protein 2 [Exaiptasia pallida] 2e-97; gi|1007745427|ref|XP_015812752.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Nothobranchius furzeri] 1e-70; gi|974077539|ref|XP_015236384.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Cyprinodon variegatus] 3e-70; gi|1129921158|ref|XP_019724841.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Hippocampus comes] 4e-70; gi|961849323|ref|XP_014842528.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia mexicana] 5e-70; gi|1143360576|ref|XP_019965131.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Paralichthys olivaceus] 1e-69; gi|961867677|ref|XP_014894293.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia latipinna] 1e-69; gi|831580677|ref|XP_012738259.1|PREDICTED: polypyrimidine tract-binding protein 2 isoform X1 [Fundulus heteroclitus] 1e-69; gi|551491094|ref|XP_005797119.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Xiphophorus maculatus] 1e-69; | ||||||
| NvERTx.4.149811 | R-1 | 0.870259999329 | E-5 | 0.999999999996 | Q66H20 | Polypyrimidine tract-binding protein 2 | gi|156392443|ref|XP_001636058.1|predicted protein [Nematostella vectensis], 8e-125 |
| other nr_hits | gi|999984893|gb|KXJ24034.1|Polypyrimidine tract-binding protein 2 [Exaiptasia pallida] 2e-97; gi|1007745427|ref|XP_015812752.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Nothobranchius furzeri] 8e-71; gi|974077539|ref|XP_015236384.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Cyprinodon variegatus] 2e-70; gi|1129921158|ref|XP_019724841.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Hippocampus comes] 3e-70; gi|961849323|ref|XP_014842528.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia mexicana] 4e-70; gi|1143360576|ref|XP_019965131.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Paralichthys olivaceus] 9e-70; gi|961867677|ref|XP_014894293.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia latipinna] 9e-70; gi|831580677|ref|XP_012738259.1|PREDICTED: polypyrimidine tract-binding protein 2 isoform X1 [Fundulus heteroclitus] 9e-70; gi|551491094|ref|XP_005797119.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Xiphophorus maculatus] 9e-70; | ||||||
| NvERTx.4.149812 | R-1 | 0.870259999329 | E-5 | 0.999999999996 | Q66H20 | Polypyrimidine tract-binding protein 2 | gi|156392443|ref|XP_001636058.1|predicted protein [Nematostella vectensis], 8e-125 |
| other nr_hits | gi|999984893|gb|KXJ24034.1|Polypyrimidine tract-binding protein 2 [Exaiptasia pallida] 2e-97; gi|1007745427|ref|XP_015812752.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Nothobranchius furzeri] 8e-71; gi|974077539|ref|XP_015236384.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Cyprinodon variegatus] 2e-70; gi|1129921158|ref|XP_019724841.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Hippocampus comes] 3e-70; gi|961849323|ref|XP_014842528.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia mexicana] 4e-70; gi|1143360576|ref|XP_019965131.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Paralichthys olivaceus] 9e-70; gi|961867677|ref|XP_014894293.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia latipinna] 9e-70; gi|831580677|ref|XP_012738259.1|PREDICTED: polypyrimidine tract-binding protein 2 isoform X1 [Fundulus heteroclitus] 9e-70; gi|551491094|ref|XP_005797119.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Xiphophorus maculatus] 9e-70; | ||||||
| NvERTx.4.149813 | R-1 | 0.870259999329 | E-5 | 0.999999999996 | Q66H20 | Polypyrimidine tract-binding protein 2 | gi|156392443|ref|XP_001636058.1|predicted protein [Nematostella vectensis], 2e-213 |
| other nr_hits | gi|999984893|gb|KXJ24034.1|Polypyrimidine tract-binding protein 2 [Exaiptasia pallida] 6e-174; gi|585653530|ref|XP_006815130.1|PREDICTED: polypyrimidine tract-binding protein 1-like isoform X2 [Saccoglossus kowalevskii] 5e-124; gi|1007745427|ref|XP_015812752.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Nothobranchius furzeri] 2e-122; gi|974077539|ref|XP_015236384.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Cyprinodon variegatus] 3e-122; gi|1129921158|ref|XP_019724841.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Hippocampus comes] 8e-122; gi|961849323|ref|XP_014842528.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia mexicana] 8e-122; gi|563317853|ref|NP_001274106.1|polypyrimidine tract binding protein 2 [Oryzias latipes] 1e-121; gi|939295055|ref|XP_012780415.2|PREDICTED: polypyrimidine tract-binding protein 2 isoform X3 [Maylandia zebra] 2e-121; gi|930756860|ref|XP_014194575.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X3 [Haplochromis burtoni] 2e-121; | ||||||
| NvERTx.4.149814 | R-1 | 0.870259999329 | E-5 | 0.999999999996 | P17225 | Polypyrimidine tract-binding protein 1 | gi|156392443|ref|XP_001636058.1|predicted protein [Nematostella vectensis], 1e-124 |
| other nr_hits | gi|999984893|gb|KXJ24034.1|Polypyrimidine tract-binding protein 2 [Exaiptasia pallida] 2e-97; gi|1007745427|ref|XP_015812752.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Nothobranchius furzeri] 1e-70; gi|974077539|ref|XP_015236384.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Cyprinodon variegatus] 3e-70; gi|1129921158|ref|XP_019724841.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Hippocampus comes] 4e-70; gi|961849323|ref|XP_014842528.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia mexicana] 5e-70; gi|1143360576|ref|XP_019965131.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Paralichthys olivaceus] 1e-69; gi|961867677|ref|XP_014894293.1|PREDICTED: polypyrimidine tract-binding protein 2-like isoform X1 [Poecilia latipinna] 1e-69; gi|831580677|ref|XP_012738259.1|PREDICTED: polypyrimidine tract-binding protein 2 isoform X1 [Fundulus heteroclitus] 1e-69; gi|551491094|ref|XP_005797119.1|PREDICTED: polypyrimidine tract-binding protein 2-like [Xiphophorus maculatus] 1e-69; | ||||||
| NvERTx.4.149816 | R-1 | 0.870259999329 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.158671 | R-3 | 0.974733494253 | E-5 | 0.999999999996 | E1C8P7 | Down syndrome cell adhesion molecule-like protein 1 homolog | gi|156391040|ref|XP_001635577.1|predicted protein [Nematostella vectensis], 1e-79 |
| other nr_hits | gi|999969067|gb|KXJ08241.1|Neural cell adhesion molecule 1 [Exaiptasia pallida] 5e-21; gi|156391044|ref|XP_001635579.1|predicted protein [Nematostella vectensis] 6e-20; gi|999984023|gb|KXJ23164.1|Neural cell adhesion molecule 1 [Exaiptasia pallida] 2e-18; gi|1005454855|ref|XP_015764570.1|PREDICTED: neural cell adhesion molecule 1-like isoform X1 [Acropora digitifera] 1e-16; gi|1005454863|ref|XP_015764574.1|PREDICTED: neural cell adhesion molecule 1-like isoform X2 [Acropora digitifera] 1e-16; gi|1005454869|ref|XP_015764577.1|PREDICTED: neural cell adhesion molecule 1-like isoform X3 [Acropora digitifera] 1e-16; gi|1005491446|ref|XP_015748397.1|PREDICTED: netrin receptor DCC-like isoform X2 [Acropora digitifera] 2e-14; gi|1005491444|ref|XP_015748396.1|PREDICTED: hemicentin-1-like isoform X1 [Acropora digitifera] 3e-14; gi|156391042|ref|XP_001635578.1|predicted protein [Nematostella vectensis] 1e-13; | ||||||
| NvERTx.4.158673 | R-3 | 0.974733494253 | E-5 | 0.999999999996 | Q8WZ42 | Titin | gi|156391040|ref|XP_001635577.1|predicted protein [Nematostella vectensis], 5e-140 |
| other nr_hits | gi|999969067|gb|KXJ08241.1|Neural cell adhesion molecule 1 [Exaiptasia pallida] 2e-36; gi|1005454855|ref|XP_015764570.1|PREDICTED: neural cell adhesion molecule 1-like isoform X1 [Acropora digitifera] 9e-32; gi|1005454863|ref|XP_015764574.1|PREDICTED: neural cell adhesion molecule 1-like isoform X2 [Acropora digitifera] 9e-32; gi|1005454869|ref|XP_015764577.1|PREDICTED: neural cell adhesion molecule 1-like isoform X3 [Acropora digitifera] 9e-32; gi|156391044|ref|XP_001635579.1|predicted protein [Nematostella vectensis] 5e-31; gi|999984023|gb|KXJ23164.1|Neural cell adhesion molecule 1 [Exaiptasia pallida] 4e-30; gi|1005491444|ref|XP_015748396.1|PREDICTED: hemicentin-1-like isoform X1 [Acropora digitifera] 3e-29; gi|1005491446|ref|XP_015748397.1|PREDICTED: netrin receptor DCC-like isoform X2 [Acropora digitifera] 7e-29; gi|156390924|ref|XP_001635519.1|predicted protein [Nematostella vectensis] 2e-19; | ||||||
| NvERTx.4.158674 | R-3 | 0.974733494253 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.158675 | R-3 | 0.974733494253 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.95586 | R-7 | 0.90776342862 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.95590 | R-7 | 0.90776342862 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.95591 | R-7 | 0.90776342862 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.95592 | R-7 | 0.90776342862 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.95593 | R-7 | 0.90776342862 | E-5 | 0.999999999996 | Q8N5I9 | Uncharacterized protein C12orf45 | gi|156400744|ref|XP_001638952.1|predicted protein [Nematostella vectensis], 4e-77 |
| other nr_hits | gi|156334780|ref|XP_001619522.1|hypothetical protein NEMVEDRAFT_v1g224097 [Nematostella vectensis] 1e-58; gi|999980879|gb|KXJ20053.1|Uncharacterized protein C12orf45 [Exaiptasia pallida] 7e-19; gi|301617369|ref|XP_002938117.1|PREDICTED: uncharacterized protein C12orf45 homolog [Xenopus tropicalis] 2e-08; gi|1072249130|ref|XP_018409383.1|PREDICTED: uncharacterized protein C12orf45 homolog [Nanorana parkeri] 9e-08; gi|291241304|ref|XP_002740554.1|PREDICTED: uncharacterized protein C12orf45 homolog [Saccoglossus kowalevskii] 1e-07; gi|432942482|ref|XP_004083007.1|PREDICTED: uncharacterized protein C12orf45 homolog [Oryzias latipes] 3e-07; gi|930751783|ref|XP_014193308.1|PREDICTED: uncharacterized protein C12orf45 homolog isoform X1 [Haplochromis burtoni] 3e-07; gi|930751785|ref|XP_014193309.1|PREDICTED: uncharacterized protein C12orf45 homolog isoform X2 [Haplochromis burtoni] 3e-07; gi|499040601|ref|XP_004570925.1|PREDICTED: uncharacterized protein C12orf45 homolog [Maylandia zebra] 3e-07; | ||||||
| NvERTx.4.95594 | R-7 | 0.90776342862 | E-5 | 0.999999999996 | No_Uniprotmatch | No_Description | gi|156400744|ref|XP_001638952.1|predicted protein [Nematostella vectensis], 1e-18 |
| other nr_hits | gi|156334780|ref|XP_001619522.1|hypothetical protein NEMVEDRAFT_v1g224097 [Nematostella vectensis] 5e-06; | ||||||
