Explore Mfuzz Clusters
Mfuzz cluster
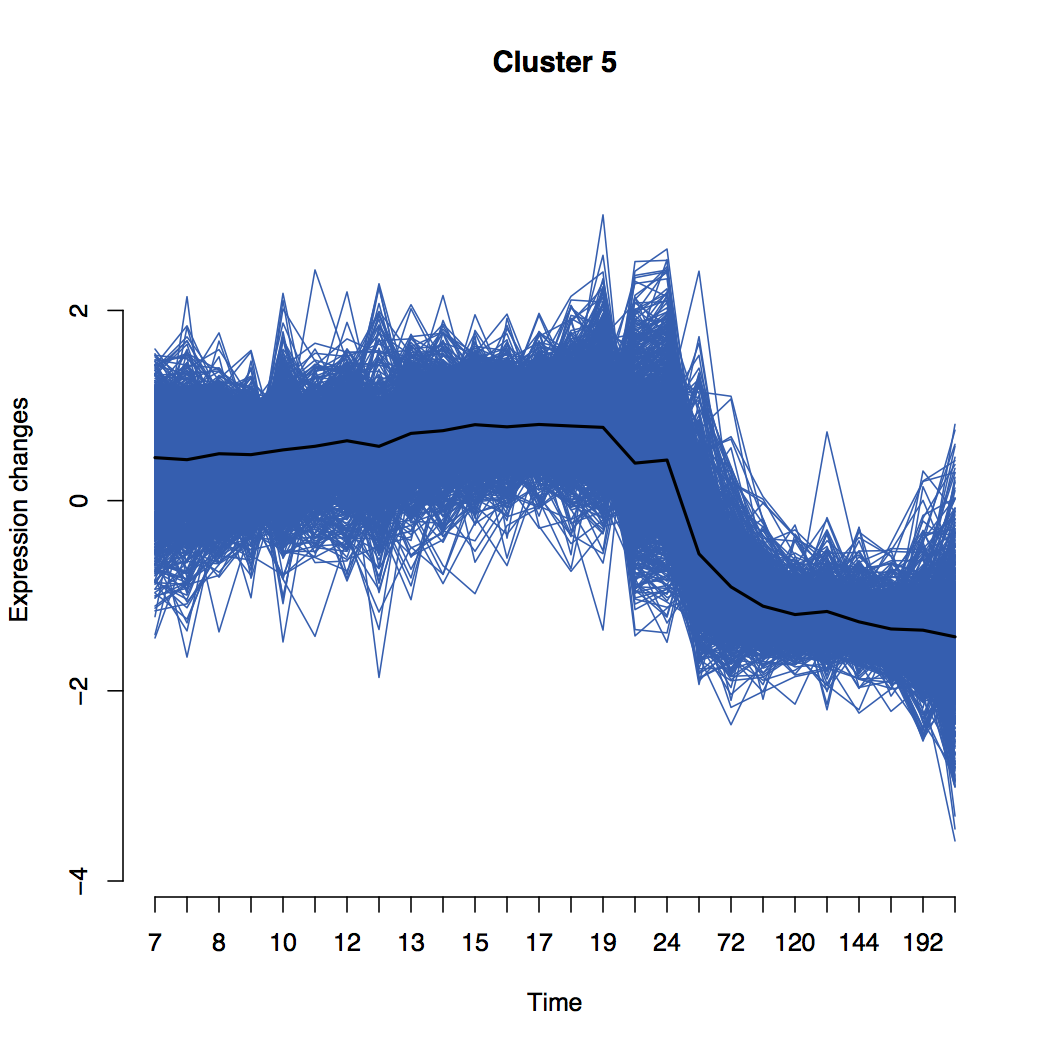
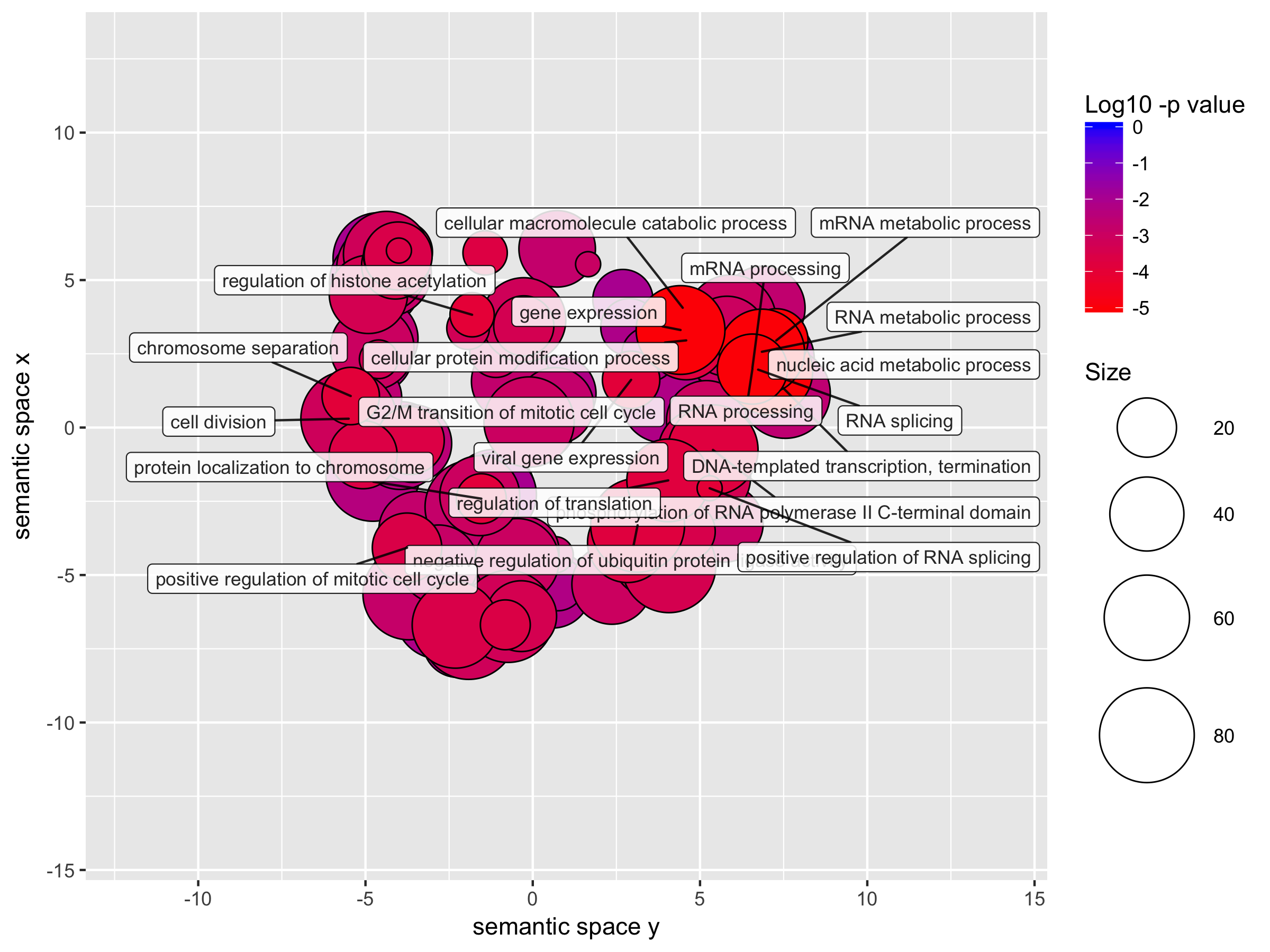
Details
click on a row to view the other nr hits
| ID | mfuzz regen clust | mfuzz regen score | mfuzz embryo clust | mfuzz embryo score | Uniprot ID | Uniprot description | top nr_hit eval |
|---|---|---|---|---|---|---|---|
| NvERTx.4.27456 | R-5 | 0.969742585068 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.27458 | R-5 | 0.969742585068 | E-5 | 1.0 | Q00899 | Transcriptional repressor protein YY1 | gi|156371552|ref|XP_001628827.1|predicted protein [Nematostella vectensis], 9e-08 |
| other nr_hits | gi|156603240|ref|XP_001618798.1|hypothetical protein NEMVEDRAFT_v1g248876 [Nematostella vectensis] 9e-08; gi|999980678|gb|KXJ19852.1|Transcriptional repressor protein YY1 [Exaiptasia pallida] 2e-07; gi|1005419159|ref|XP_015751286.1|PREDICTED: LOW QUALITY PROTEIN: transcriptional repressor protein YY1-like [Acropora digitifera] 3e-06; | ||||||
| NvERTx.4.55019 | R-5 | 0.969742585068 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.66716 | R-5 | 0.969742585068 | E-5 | 1.0 | P25490 | Transcriptional repressor protein YY1 | gi|999980678|gb|KXJ19852.1|Transcriptional repressor protein YY1 [Exaiptasia pallida], 8e-122 |
| other nr_hits | gi|1005419159|ref|XP_015751286.1|PREDICTED: LOW QUALITY PROTEIN: transcriptional repressor protein YY1-like [Acropora digitifera] 2e-103; gi|1126254325|ref|XP_019653320.1|PREDICTED: transcriptional repressor protein YY1 [Ailuropoda melanoleuca] 5e-90; gi|657734119|ref|XP_008305638.1|PREDICTED: transcriptional repressor protein YY1-like [Cynoglossus semilaevis] 5e-90; gi|1121913850|ref|XP_019408054.1|PREDICTED: transcriptional repressor protein YY1 [Crocodylus porosus] 3e-89; gi|1003813528|ref|XP_015721123.1|PREDICTED: transcriptional repressor protein YY1 [Coturnix japonica] 3e-89; gi|998697941|ref|XP_015486582.1|PREDICTED: transcriptional repressor protein YY1 isoform X1 [Parus major] 3e-89; gi|950951757|ref|XP_014451969.1|PREDICTED: transcriptional repressor protein YY1 [Alligator mississippiensis] 3e-89; gi|960994973|ref|XP_014813043.1|PREDICTED: transcriptional repressor protein YY1 [Calidris pugnax] 3e-89; gi|929445844|ref|XP_014135152.1|PREDICTED: transcriptional repressor protein YY1 [Falco cherrug] 3e-89; | ||||||
| NvERTx.4.67997 | R-5 | 0.999698373287 | E-5 | 1.0 | Q8C9M2 | Coiled-coil domain-containing protein 15 | gi|156402860|ref|XP_001639808.1|predicted protein [Nematostella vectensis], 8e-279 |
| other nr_hits | gi|999989754|gb|KXJ28786.1|Coiled-coil domain-containing protein 15 [Exaiptasia pallida] 9e-100; gi|1005453580|ref|XP_015763951.1|PREDICTED: uncharacterized protein LOC107342946 [Acropora digitifera] 3e-94; gi|585703670|ref|XP_002740124.2|PREDICTED: coiled-coil domain-containing protein 15-like [Saccoglossus kowalevskii] 1e-70; gi|556961939|ref|XP_005991201.1|PREDICTED: coiled-coil domain-containing protein 15 isoform X1 [Latimeria chalumnae] 3e-51; gi|196012862|ref|XP_002116293.1|hypothetical protein TRIADDRAFT_60238 [Trichoplax adhaerens] 1e-50; gi|443720216|gb|ELU10015.1|hypothetical protein CAPTEDRAFT_221405 [Capitella teleta] 2e-50; gi|556961946|ref|XP_005991203.1|PREDICTED: coiled-coil domain-containing protein 15 isoform X2 [Latimeria chalumnae] 4e-49; gi|147907024|ref|NP_001090154.1|coiled-coil domain containing 15 S homeolog [Xenopus laevis] 7e-49; gi|871204283|ref|XP_005088844.2|PREDICTED: coiled-coil domain-containing protein 15-like [Aplysia californica] 9e-49; | ||||||
| NvERTx.4.33560 | R-5 | 0.481267171772 | E-5 | 1.0 | Q6GLD8 | Cyclin-dependent kinase 9 | gi|156365870|ref|XP_001626865.1|predicted protein [Nematostella vectensis], 3e-200 |
| other nr_hits | gi|999970690|gb|KXJ09864.1|Cyclin-dependent kinase 9 [Exaiptasia pallida] 5e-175; gi|1005445772|ref|XP_015760180.1|PREDICTED: cyclin-dependent kinase 9-like isoform X1 [Acropora digitifera] 3e-167; gi|926617272|ref|XP_013773793.1|PREDICTED: cyclin-dependent kinase 9-like [Limulus polyphemus] 3e-152; gi|530600025|ref|XP_005293504.1|PREDICTED: cyclin-dependent kinase 9 [Chrysemys picta bellii] 4e-149; gi|585667589|ref|XP_006817341.1|PREDICTED: cyclin-dependent kinase 9-like [Saccoglossus kowalevskii] 6e-149; gi|1058218950|gb|JAT12991.1|hypothetical protein g.29115 [Graphocephala atropunctata] 1e-148; gi|1051166546|ref|XP_017665897.1|PREDICTED: cyclin-dependent kinase 9 [Lepidothrix coronata] 1e-148; gi|525015929|ref|XP_005055538.1|PREDICTED: cyclin-dependent kinase 9 [Ficedula albicollis] 1e-148; gi|847153765|ref|XP_012823647.1|PREDICTED: cyclin-dependent kinase 9 isoform X2 [Xenopus tropicalis] 2e-148; | ||||||
| NvERTx.4.33561 | R-5 | 0.481267171772 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.33562 | R-5 | 0.481267171772 | E-5 | 1.0 | Q5ZKN1 | Cyclin-dependent kinase 9 | gi|156365870|ref|XP_001626865.1|predicted protein [Nematostella vectensis], 4e-170 |
| other nr_hits | gi|999970690|gb|KXJ09864.1|Cyclin-dependent kinase 9 [Exaiptasia pallida] 2e-149; gi|1005445772|ref|XP_015760180.1|PREDICTED: cyclin-dependent kinase 9-like isoform X1 [Acropora digitifera] 3e-142; gi|1005445778|ref|XP_015760184.1|PREDICTED: cyclin-dependent kinase 9-like isoform X4 [Acropora digitifera] 3e-142; gi|1005445774|ref|XP_015760181.1|PREDICTED: cyclin-dependent kinase 9-like isoform X2 [Acropora digitifera] 8e-138; gi|1005445776|ref|XP_015760183.1|PREDICTED: cyclin-dependent kinase 9-like isoform X3 [Acropora digitifera] 8e-138; gi|926617272|ref|XP_013773793.1|PREDICTED: cyclin-dependent kinase 9-like [Limulus polyphemus] 2e-132; gi|930234828|ref|XP_005421475.2|PREDICTED: cyclin-dependent kinase 9 [Geospiza fortis] 4e-131; gi|602657598|ref|XP_007434515.1|PREDICTED: cyclin-dependent kinase 9 [Python bivittatus] 1e-130; gi|704173416|ref|XP_010132656.1|PREDICTED: cyclin-dependent kinase 9, partial [Buceros rhinoceros silvestris] 2e-130; | ||||||
| NvERTx.4.33563 | R-5 | 0.481267171772 | E-5 | 1.0 | Q6GLD8 | Cyclin-dependent kinase 9 | gi|156365870|ref|XP_001626865.1|predicted protein [Nematostella vectensis], 1e-24 |
| other nr_hits | gi|999970690|gb|KXJ09864.1|Cyclin-dependent kinase 9 [Exaiptasia pallida] 3e-20; gi|1005445772|ref|XP_015760180.1|PREDICTED: cyclin-dependent kinase 9-like isoform X1 [Acropora digitifera] 1e-19; gi|732618161|gb|JAG24504.1|Cyclin-dependent kinase 9 [Lygus hesperus] 9e-16; gi|1009351175|gb|KYM76659.1|Cell division protein kinase 9 [Atta colombica] 9e-16; gi|965555432|gb|JAP03579.1|putative cdc2-related protein kinase [Triatoma dimidiata] 2e-15; gi|656764487|gb|JAC87117.1|putative cdc2-related protein kinase [Panstrongylus megistus] 2e-15; gi|604791212|gb|JAC18052.1|putative cdc2-related protein kinase [Triatoma infestans] 2e-15; gi|1036979259|ref|XP_017148178.1|PREDICTED: cyclin-dependent kinase 9 [Drosophila miranda] 2e-15; gi|195151075|ref|XP_002016473.1|GL10467 [Drosophila persimilis] 2e-15; | ||||||
| NvERTx.4.88129 | R-5 | 0.999955457804 | E-5 | 1.0 | Q5ZJ24 | LisH domain-containing protein FOPNL | gi|156386359|ref|XP_001633880.1|predicted protein [Nematostella vectensis], 2e-84 |
| other nr_hits | gi|156344489|ref|XP_001621204.1|hypothetical protein NEMVEDRAFT_v1g4745 [Nematostella vectensis] 2e-53; gi|291226611|ref|XP_002733287.1|PREDICTED: lisH domain-containing protein FOPNL-like [Saccoglossus kowalevskii] 4e-40; gi|1005462816|ref|XP_015768444.1|PREDICTED: lisH domain-containing protein FOPNL-like isoform X1 [Acropora digitifera] 8e-39; gi|573899291|ref|XP_006637377.1|PREDICTED: lisH domain-containing protein FOPNL isoform X2 [Lepisosteus oculatus] 1e-38; gi|762170186|ref|XP_011423515.1|PREDICTED: lisH domain-containing protein FOPNL [Crassostrea gigas] 1e-37; gi|405965776|gb|EKC31130.1|LisH domain-containing protein C16orf63-like protein [Crassostrea gigas] 1e-37; gi|727001233|ref|XP_010390815.1|PREDICTED: lisH domain-containing protein FOPNL isoform X4 [Corvus cornix cornix] 2e-37; gi|390333747|ref|XP_781294.2|PREDICTED: lisH domain-containing protein FOPNL [Strongylocentrotus purpuratus] 3e-37; gi|959036661|ref|XP_014750849.1|PREDICTED: lisH domain-containing protein FOPNL isoform X2 [Sturnus vulgaris] 4e-37; | ||||||
| NvERTx.4.88130 | R-5 | 0.999955457804 | E-5 | 1.0 | Q5ZJ24 | LisH domain-containing protein FOPNL | gi|156386359|ref|XP_001633880.1|predicted protein [Nematostella vectensis], 3e-50 |
| other nr_hits | gi|156344489|ref|XP_001621204.1|hypothetical protein NEMVEDRAFT_v1g4745 [Nematostella vectensis] 6e-45; gi|291226611|ref|XP_002733287.1|PREDICTED: lisH domain-containing protein FOPNL-like [Saccoglossus kowalevskii] 3e-34; gi|762170186|ref|XP_011423515.1|PREDICTED: lisH domain-containing protein FOPNL [Crassostrea gigas] 6e-34; gi|405965776|gb|EKC31130.1|LisH domain-containing protein C16orf63-like protein [Crassostrea gigas] 6e-34; gi|1005462816|ref|XP_015768444.1|PREDICTED: lisH domain-containing protein FOPNL-like isoform X1 [Acropora digitifera] 4e-33; gi|919016153|ref|XP_013392432.1|PREDICTED: lisH domain-containing protein FOPNL-like [Lingula anatina] 2e-32; gi|727001231|ref|XP_010390813.1|PREDICTED: lisH domain-containing protein FOPNL isoform X3 [Corvus cornix cornix] 4e-32; gi|727001233|ref|XP_010390815.1|PREDICTED: lisH domain-containing protein FOPNL isoform X4 [Corvus cornix cornix] 4e-32; gi|390333747|ref|XP_781294.2|PREDICTED: lisH domain-containing protein FOPNL [Strongylocentrotus purpuratus] 6e-32; | ||||||
| NvERTx.4.88131 | R-5 | 0.999955457804 | E-5 | 1.0 | Q5ZJ24 | LisH domain-containing protein FOPNL | gi|156386359|ref|XP_001633880.1|predicted protein [Nematostella vectensis], 3e-50 |
| other nr_hits | gi|156344489|ref|XP_001621204.1|hypothetical protein NEMVEDRAFT_v1g4745 [Nematostella vectensis] 6e-45; gi|291226611|ref|XP_002733287.1|PREDICTED: lisH domain-containing protein FOPNL-like [Saccoglossus kowalevskii] 3e-34; gi|762170186|ref|XP_011423515.1|PREDICTED: lisH domain-containing protein FOPNL [Crassostrea gigas] 6e-34; gi|405965776|gb|EKC31130.1|LisH domain-containing protein C16orf63-like protein [Crassostrea gigas] 6e-34; gi|1005462816|ref|XP_015768444.1|PREDICTED: lisH domain-containing protein FOPNL-like isoform X1 [Acropora digitifera] 4e-33; gi|919016153|ref|XP_013392432.1|PREDICTED: lisH domain-containing protein FOPNL-like [Lingula anatina] 2e-32; gi|727001231|ref|XP_010390813.1|PREDICTED: lisH domain-containing protein FOPNL isoform X3 [Corvus cornix cornix] 4e-32; gi|727001233|ref|XP_010390815.1|PREDICTED: lisH domain-containing protein FOPNL isoform X4 [Corvus cornix cornix] 4e-32; gi|390333747|ref|XP_781294.2|PREDICTED: lisH domain-containing protein FOPNL [Strongylocentrotus purpuratus] 6e-32; | ||||||
| NvERTx.4.88132 | R-5 | 0.999955457804 | E-5 | 1.0 | Q5ZJ24 | LisH domain-containing protein FOPNL | gi|156386359|ref|XP_001633880.1|predicted protein [Nematostella vectensis], 7e-83 |
| other nr_hits | gi|156344489|ref|XP_001621204.1|hypothetical protein NEMVEDRAFT_v1g4745 [Nematostella vectensis] 1e-53; gi|291226611|ref|XP_002733287.1|PREDICTED: lisH domain-containing protein FOPNL-like [Saccoglossus kowalevskii] 7e-40; gi|1005462816|ref|XP_015768444.1|PREDICTED: lisH domain-containing protein FOPNL-like isoform X1 [Acropora digitifera] 1e-38; gi|573899291|ref|XP_006637377.1|PREDICTED: lisH domain-containing protein FOPNL isoform X2 [Lepisosteus oculatus] 2e-38; gi|762170186|ref|XP_011423515.1|PREDICTED: lisH domain-containing protein FOPNL [Crassostrea gigas] 2e-37; gi|405965776|gb|EKC31130.1|LisH domain-containing protein C16orf63-like protein [Crassostrea gigas] 2e-37; gi|390333747|ref|XP_781294.2|PREDICTED: lisH domain-containing protein FOPNL [Strongylocentrotus purpuratus] 5e-37; gi|1079790580|ref|XP_018520363.1|PREDICTED: lisH domain-containing protein FOPNL [Lates calcarifer] 9e-37; gi|727001233|ref|XP_010390815.1|PREDICTED: lisH domain-containing protein FOPNL isoform X4 [Corvus cornix cornix] 9e-37; | ||||||
| NvERTx.4.84629 | R-5 | 0.618793814454 | E-5 | 1.0 | Q8CIG3 | Lysine-specific histone demethylase 1B | gi|999975736|gb|KXJ14910.1|Lysine-specific histone demethylase 1B [Exaiptasia pallida], 5e-59 |
| other nr_hits | gi|1126199251|ref|XP_019632884.1|PREDICTED: lysine-specific histone demethylase 1B-like [Branchiostoma belcheri] 7e-40; gi|612008187|ref|XP_007487955.1|PREDICTED: lysine-specific histone demethylase 1B [Monodelphis domestica] 3e-36; gi|1020567998|ref|XP_016113073.1|PREDICTED: lysine-specific histone demethylase 1B [Sinocyclocheilus grahami] 8e-36; gi|1025387584|ref|XP_016404297.1|PREDICTED: lysine-specific histone demethylase 1B [Sinocyclocheilus rhinocerous] 1e-35; gi|1101575404|ref|XP_018953388.1|PREDICTED: lysine-specific histone demethylase 1B-like, partial [Cyprinus carpio] 2e-35; gi|260788151|ref|XP_002589114.1|hypothetical protein BRAFLDRAFT_213824 [Branchiostoma floridae] 2e-35; gi|831312610|ref|XP_012688807.1|PREDICTED: lysine-specific histone demethylase 1B [Clupea harengus] 3e-35; gi|821465564|ref|XP_012397067.1|PREDICTED: LOW QUALITY PROTEIN: lysine-specific histone demethylase 1B [Sarcophilus harrisii] 4e-35; gi|1121802988|ref|XP_019399421.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Crocodylus porosus] 1e-34; | ||||||
| NvERTx.4.84630 | R-5 | 0.618793814454 | E-5 | 1.0 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.84631 | R-5 | 0.618793814454 | E-5 | 1.0 | Q8NB78 | Lysine-specific histone demethylase 1B | gi|1126199251|ref|XP_019632884.1|PREDICTED: lysine-specific histone demethylase 1B-like [Branchiostoma belcheri], 1e-204 |
| other nr_hits | gi|677298076|gb|KFP80504.1|Lysine-specific histone demethylase 1B [Apaloderma vittatum] 1e-197; gi|327270106|ref|XP_003219832.1|PREDICTED: lysine-specific histone demethylase 1B [Anolis carolinensis] 3e-197; gi|557289313|ref|XP_006027302.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Alligator sinensis] 4e-197; gi|612008187|ref|XP_007487955.1|PREDICTED: lysine-specific histone demethylase 1B [Monodelphis domestica] 4e-197; gi|1117244967|ref|XP_019378801.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Gavialis gangeticus] 6e-197; gi|830197441|ref|XP_012588410.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Condylura cristata] 8e-197; gi|704471571|ref|XP_010071246.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Pterocles gutturalis] 1e-196; gi|697831738|ref|XP_009636233.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Egretta garzetta] 1e-196; gi|1121802988|ref|XP_019399421.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Crocodylus porosus] 2e-196; | ||||||
| NvERTx.4.84632 | R-5 | 0.618793814454 | E-5 | 1.0 | Q8CIG3 | Lysine-specific histone demethylase 1B | gi|999975736|gb|KXJ14910.1|Lysine-specific histone demethylase 1B [Exaiptasia pallida], 1e-93 |
| other nr_hits | gi|1126199251|ref|XP_019632884.1|PREDICTED: lysine-specific histone demethylase 1B-like [Branchiostoma belcheri] 2e-60; gi|919034718|ref|XP_013400926.1|PREDICTED: lysine-specific histone demethylase 1B-like [Lingula anatina] 1e-55; gi|1005430243|ref|XP_015752864.1|PREDICTED: lysine-specific histone demethylase 1B-like [Acropora digitifera] 1e-55; gi|780035433|ref|XP_011667313.1|PREDICTED: lysine-specific histone demethylase 1B [Strongylocentrotus purpuratus] 6e-55; gi|1101575404|ref|XP_018953388.1|PREDICTED: lysine-specific histone demethylase 1B-like, partial [Cyprinus carpio] 5e-54; gi|926636643|ref|XP_013784265.1|PREDICTED: lysine-specific histone demethylase 1B-like, partial [Limulus polyphemus] 6e-54; gi|612008187|ref|XP_007487955.1|PREDICTED: lysine-specific histone demethylase 1B [Monodelphis domestica] 8e-54; gi|1025387584|ref|XP_016404297.1|PREDICTED: lysine-specific histone demethylase 1B [Sinocyclocheilus rhinocerous] 1e-53; gi|765134844|ref|XP_011479325.1|PREDICTED: lysine-specific histone demethylase 1B [Oryzias latipes] 2e-53; | ||||||
| NvERTx.4.84633 | R-5 | 0.618793814454 | E-5 | 1.0 | Q61184 | Melatonin receptor type 1A | gi|1005487077|ref|XP_015780129.1|PREDICTED: visual pigment-like receptor peropsin [Acropora digitifera], 2e-29 |
| other nr_hits | gi|1005447617|ref|XP_015761092.1|PREDICTED: melatonin receptor type 1A-like [Acropora digitifera] 5e-26; gi|156378530|ref|XP_001631195.1|predicted protein [Nematostella vectensis] 4e-24; gi|1005465494|ref|XP_015769737.1|PREDICTED: melatonin-related receptor-like [Acropora digitifera] 8e-24; gi|1005441220|ref|XP_015757948.1|PREDICTED: muscarinic acetylcholine receptor M1-like [Acropora digitifera] 1e-23; gi|156408089|ref|XP_001641689.1|predicted protein [Nematostella vectensis] 3e-23; gi|156378588|ref|XP_001631224.1|predicted protein [Nematostella vectensis] 7e-22; gi|1005478918|ref|XP_015776173.1|PREDICTED: melatonin receptor type 1A-like [Acropora digitifera] 1e-21; gi|1005488962|ref|XP_015747190.1|PREDICTED: adenosine receptor A3-like [Acropora digitifera] 1e-21; gi|156378532|ref|XP_001631196.1|predicted protein [Nematostella vectensis] 2e-21; | ||||||
| NvERTx.4.84634 | R-5 | 0.618793814454 | E-5 | 1.0 | Q8NB78 | Lysine-specific histone demethylase 1B | gi|1005430243|ref|XP_015752864.1|PREDICTED: lysine-specific histone demethylase 1B-like [Acropora digitifera], 5e-122 |
| other nr_hits | gi|1126199251|ref|XP_019632884.1|PREDICTED: lysine-specific histone demethylase 1B-like [Branchiostoma belcheri] 4e-119; gi|677298076|gb|KFP80504.1|Lysine-specific histone demethylase 1B [Apaloderma vittatum] 5e-118; gi|830197441|ref|XP_012588410.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Condylura cristata] 7e-118; gi|821130568|ref|XP_012386324.1|PREDICTED: lysine-specific histone demethylase 1B [Dasypus novemcinctus] 7e-118; gi|803221834|ref|XP_011971217.1|PREDICTED: lysine-specific histone demethylase 1B [Ovis aries musimon] 7e-118; gi|1062951632|ref|XP_017894670.1|PREDICTED: lysine-specific histone demethylase 1B [Capra hircus] 7e-118; gi|965993645|ref|XP_014958315.1|PREDICTED: lysine-specific histone demethylase 1B [Ovis aries] 7e-118; gi|677366350|gb|KFP94988.1|Lysine-specific histone demethylase 1B, partial [Haliaeetus albicilla] 1e-117; gi|704471571|ref|XP_010071246.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Pterocles gutturalis] 1e-117; | ||||||
| NvERTx.4.84635 | R-5 | 0.618793814454 | E-5 | 1.0 | Q8CIG3 | Lysine-specific histone demethylase 1B | gi|999975736|gb|KXJ14910.1|Lysine-specific histone demethylase 1B [Exaiptasia pallida], 5e-119 |
| other nr_hits | gi|1126199251|ref|XP_019632884.1|PREDICTED: lysine-specific histone demethylase 1B-like [Branchiostoma belcheri] 1e-76; gi|1005430243|ref|XP_015752864.1|PREDICTED: lysine-specific histone demethylase 1B-like [Acropora digitifera] 4e-75; gi|612008187|ref|XP_007487955.1|PREDICTED: lysine-specific histone demethylase 1B [Monodelphis domestica] 1e-71; gi|327270106|ref|XP_003219832.1|PREDICTED: lysine-specific histone demethylase 1B [Anolis carolinensis] 2e-71; gi|944346219|ref|XP_014377087.1|PREDICTED: lysine-specific histone demethylase 1B isoform X2 [Alligator sinensis] 3e-71; gi|821465564|ref|XP_012397067.1|PREDICTED: LOW QUALITY PROTEIN: lysine-specific histone demethylase 1B [Sarcophilus harrisii] 3e-71; gi|557289313|ref|XP_006027302.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Alligator sinensis] 3e-71; gi|1121802988|ref|XP_019399421.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Crocodylus porosus] 4e-71; gi|1121802990|ref|XP_019399429.1|PREDICTED: lysine-specific histone demethylase 1B isoform X2 [Crocodylus porosus] 4e-71; | ||||||
| NvERTx.4.84636 | R-5 | 0.618793814454 | E-5 | 1.0 | Q8NB78 | Lysine-specific histone demethylase 1B | gi|1005430243|ref|XP_015752864.1|PREDICTED: lysine-specific histone demethylase 1B-like [Acropora digitifera], 1e-136 |
| other nr_hits | gi|677298076|gb|KFP80504.1|Lysine-specific histone demethylase 1B [Apaloderma vittatum] 8e-129; gi|704471571|ref|XP_010071246.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Pterocles gutturalis] 7e-128; gi|701318095|ref|XP_010019003.1|PREDICTED: lysine-specific histone demethylase 1B, partial [Nestor notabilis] 9e-128; gi|677465830|gb|KFQ55244.1|Lysine-specific histone demethylase 1B, partial [Nestor notabilis] 9e-128; gi|697831738|ref|XP_009636233.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Egretta garzetta] 9e-128; gi|694653538|ref|XP_009483101.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Pelecanus crispus] 1e-127; gi|700394365|ref|XP_009937712.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Opisthocomus hoazin] 2e-127; gi|686592200|ref|XP_009271997.1|PREDICTED: lysine-specific histone demethylase 1B [Aptenodytes forsteri] 2e-127; gi|803221834|ref|XP_011971217.1|PREDICTED: lysine-specific histone demethylase 1B [Ovis aries musimon] 2e-127; | ||||||
| NvERTx.4.84638 | R-5 | 0.618793814454 | E-5 | 1.0 | Q8CIG3 | Lysine-specific histone demethylase 1B | gi|999975736|gb|KXJ14910.1|Lysine-specific histone demethylase 1B [Exaiptasia pallida], 3e-194 |
| other nr_hits | gi|1126199251|ref|XP_019632884.1|PREDICTED: lysine-specific histone demethylase 1B-like [Branchiostoma belcheri] 7e-166; gi|612008187|ref|XP_007487955.1|PREDICTED: lysine-specific histone demethylase 1B [Monodelphis domestica] 9e-159; gi|909794279|ref|XP_013153306.1|PREDICTED: lysine-specific histone demethylase 1B isoform X3 [Falco peregrinus] 3e-158; gi|529431961|ref|XP_005235777.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Falco peregrinus] 3e-158; gi|701358732|ref|XP_009961125.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Tyto alba] 4e-158; gi|677298076|gb|KFP80504.1|Lysine-specific histone demethylase 1B [Apaloderma vittatum] 1e-157; gi|960963368|ref|XP_014796421.1|PREDICTED: lysine-specific histone demethylase 1B [Calidris pugnax] 1e-157; gi|678195150|gb|KFV62132.1|Lysine-specific histone demethylase 1B [Picoides pubescens] 1e-157; gi|700394365|ref|XP_009937712.1|PREDICTED: lysine-specific histone demethylase 1B isoform X1 [Opisthocomus hoazin] 2e-157; | ||||||
| NvERTx.4.131859 | R-5 | 0.50724441418 | E-5 | 1.0 | Q9Z0W3 | Nuclear pore complex protein Nup160 | gi|156406038|ref|XP_001641038.1|predicted protein [Nematostella vectensis], 4e-68 |
| other nr_hits | gi|999981607|gb|KXJ20781.1|Nuclear pore complex protein Nup160 [Exaiptasia pallida] 4e-54; gi|919089931|ref|XP_013382002.1|PREDICTED: nuclear pore complex protein Nup160-like [Lingula anatina] 4e-28; gi|919017641|ref|XP_013393121.1|PREDICTED: nuclear pore complex protein Nup160-like isoform X1 [Lingula anatina] 1e-27; gi|919017644|ref|XP_013393122.1|PREDICTED: nuclear pore complex protein Nup160-like isoform X2 [Lingula anatina] 1e-27; gi|443683668|gb|ELT87837.1|hypothetical protein CAPTEDRAFT_178964 [Capitella teleta] 4e-27; gi|926633895|ref|XP_013782773.1|PREDICTED: nuclear pore complex protein Nup160-like [Limulus polyphemus] 5e-26; gi|998521305|ref|XP_015521672.1|PREDICTED: nuclear pore complex protein Nup160 homolog [Neodiprion lecontei] 3e-25; gi|524869912|ref|XP_005091744.1|PREDICTED: nuclear pore complex protein Nup160-like [Aplysia californica] 3e-25; gi|908476164|ref|XP_013090309.1|PREDICTED: nuclear pore complex protein Nup160-like [Biomphalaria glabrata] 5e-25; | ||||||
| NvERTx.4.131861 | R-5 | 0.50724441418 | E-5 | 1.0 | Q9Z0W3 | Nuclear pore complex protein Nup160 | gi|999981607|gb|KXJ20781.1|Nuclear pore complex protein Nup160 [Exaiptasia pallida], 3e-156 |
| other nr_hits | gi|919089931|ref|XP_013382002.1|PREDICTED: nuclear pore complex protein Nup160-like [Lingula anatina] 2e-102; gi|1126187099|ref|XP_019626295.1|PREDICTED: nuclear pore complex protein Nup160-like isoform X1 [Branchiostoma belcheri] 5e-85; gi|1126187103|ref|XP_019626298.1|PREDICTED: nuclear pore complex protein Nup160-like isoform X3 [Branchiostoma belcheri] 5e-85; gi|1126187105|ref|XP_019626299.1|PREDICTED: nuclear pore complex protein Nup160-like isoform X4 [Branchiostoma belcheri] 5e-85; gi|1005448102|ref|XP_015761329.1|PREDICTED: nuclear pore complex protein Nup160-like [Acropora digitifera] 1e-84; gi|926633895|ref|XP_013782773.1|PREDICTED: nuclear pore complex protein Nup160-like [Limulus polyphemus] 3e-84; gi|1139827264|ref|XP_011446987.2|PREDICTED: nuclear pore complex protein Nup160 [Crassostrea gigas] 5e-82; gi|443683668|gb|ELT87837.1|hypothetical protein CAPTEDRAFT_178964 [Capitella teleta] 5e-82; gi|908476164|ref|XP_013090309.1|PREDICTED: nuclear pore complex protein Nup160-like [Biomphalaria glabrata] 4e-80; | ||||||
| NvERTx.4.21164 | R-8 | 0.996098340069 | E-5 | 0.999999999999 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.21165 | R-8 | 0.996098340069 | E-5 | 0.999999999999 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.157872 | R-8 | 0.996098340069 | E-5 | 0.999999999999 | Q67XX3 | F-box protein At5g06550 | gi|156386727|ref|XP_001634063.1|predicted protein [Nematostella vectensis], 1e-147 |
| other nr_hits | gi|999985071|gb|KXJ24212.1|F-box protein [Exaiptasia pallida] 2e-125; gi|1126193475|ref|XP_019629782.1|PREDICTED: uncharacterized protein LOC109474018 [Branchiostoma belcheri] 1e-98; gi|1126170313|ref|XP_019617146.1|PREDICTED: uncharacterized protein LOC109464580 [Branchiostoma belcheri] 6e-95; gi|919101438|ref|XP_013387081.1|PREDICTED: uncharacterized protein LOC106156398 [Lingula anatina] 7e-95; gi|957819473|ref|XP_014674984.1|PREDICTED: uncharacterized protein LOC106815073 [Priapulus caudatus] 2e-94; gi|291232509|ref|XP_002736198.1|PREDICTED: uncharacterized protein LOC100369871 [Saccoglossus kowalevskii] 9e-93; gi|1005424945|ref|XP_015778627.1|PREDICTED: uncharacterized protein LOC107356535 [Acropora digitifera] 1e-90; gi|1067101562|ref|XP_018012909.1|PREDICTED: uncharacterized protein LOC108669977 [Hyalella azteca] 4e-90; gi|443721977|gb|ELU11050.1|hypothetical protein CAPTEDRAFT_142276 [Capitella teleta] 1e-89; | ||||||
| NvERTx.4.157873 | R-8 | 0.996098340069 | E-5 | 0.999999999999 | A0A1S3HNH6_LINUN | uncharacterized protein LOC106156398 {ECO:0000313|RefSeq:XP_013387081.1} | gi|156386727|ref|XP_001634063.1|predicted protein [Nematostella vectensis], 9e-73 |
| other nr_hits | gi|999985071|gb|KXJ24212.1|F-box protein [Exaiptasia pallida] 7e-62; gi|1005424945|ref|XP_015778627.1|PREDICTED: uncharacterized protein LOC107356535 [Acropora digitifera] 2e-55; gi|1126193475|ref|XP_019629782.1|PREDICTED: uncharacterized protein LOC109474018 [Branchiostoma belcheri] 6e-40; gi|919101438|ref|XP_013387081.1|PREDICTED: uncharacterized protein LOC106156398 [Lingula anatina] 1e-39; gi|260816163|ref|XP_002602841.1|hypothetical protein BRAFLDRAFT_128934 [Branchiostoma floridae] 2e-38; gi|957819473|ref|XP_014674984.1|PREDICTED: uncharacterized protein LOC106815073 [Priapulus caudatus] 7e-38; gi|1126170313|ref|XP_019617146.1|PREDICTED: uncharacterized protein LOC109464580 [Branchiostoma belcheri] 1e-37; gi|291232509|ref|XP_002736198.1|PREDICTED: uncharacterized protein LOC100369871 [Saccoglossus kowalevskii] 6e-37; gi|196003336|ref|XP_002111535.1|expressed hypothetical protein [Trichoplax adhaerens] 3e-34; | ||||||
| NvERTx.4.157882 | R-8 | 0.996098340069 | E-5 | 0.999999999999 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.88135 | R-7 | 0.9998327724 | E-5 | 0.999999999999 | Q8R332 | Nucleoporin p58/p45 {ECO:0000305} | gi|999984826|gb|KXJ23967.1|Nucleoporin p58/p45 [Exaiptasia pallida], 1e-84 |
| other nr_hits | gi|156365715|ref|XP_001626789.1|predicted protein [Nematostella vectensis] 3e-72; gi|919083271|ref|XP_013378988.1|PREDICTED: nucleoporin p58/p45-like isoform X1 [Lingula anatina] 4e-58; gi|919083274|ref|XP_013378989.1|PREDICTED: nucleoporin p58/p45-like isoform X2 [Lingula anatina] 4e-58; gi|1139831278|ref|XP_019929192.1|PREDICTED: probable nucleoporin Nup58 [Crassostrea gigas] 3e-55; gi|1003735484|ref|XP_015707588.1|PREDICTED: nucleoporin p58/p45 isoform X1 [Coturnix japonica] 2e-52; gi|1003735492|ref|XP_015707589.1|PREDICTED: nucleoporin p58/p45 isoform X2 [Coturnix japonica] 2e-52; gi|1003735498|ref|XP_015707590.1|PREDICTED: nucleoporin p58/p45 isoform X3 [Coturnix japonica] 2e-52; gi|971377980|ref|XP_015133489.1|PREDICTED: nucleoporin p58/p45 isoform X1 [Gallus gallus] 2e-52; gi|971377982|ref|XP_015133493.1|PREDICTED: nucleoporin p58/p45 isoform X2 [Gallus gallus] 2e-52; | ||||||
| NvERTx.4.88136 | R-7 | 0.9998327724 | E-5 | 0.999999999999 | Q8R332 | Nucleoporin p58/p45 {ECO:0000305} | gi|156365715|ref|XP_001626789.1|predicted protein [Nematostella vectensis], 1e-72 |
| other nr_hits | gi|999984826|gb|KXJ23967.1|Nucleoporin p58/p45 [Exaiptasia pallida] 1e-60; gi|1005481535|ref|XP_015777472.1|PREDICTED: nucleoporin p58/p45-like [Acropora digitifera] 1e-51; gi|537241758|gb|ERE87297.1|nucleoporin [Cricetulus griseus] 6e-37; gi|537241761|gb|ERE87300.1|nucleoporin [Cricetulus griseus] 6e-37; gi|975118868|ref|XP_015274854.1|PREDICTED: nucleoporin p58/p45 isoform X2 [Gekko japonicus] 2e-36; gi|657741217|ref|XP_008305689.1|PREDICTED: nucleoporin p58/p45 isoform X1 [Cynoglossus semilaevis] 2e-36; gi|585712152|ref|XP_006824869.1|PREDICTED: nucleoporin p58/p45-like [Saccoglossus kowalevskii] 4e-36; gi|260833598|ref|XP_002611744.1|hypothetical protein BRAFLDRAFT_128724 [Branchiostoma floridae] 5e-36; gi|972958899|ref|XP_015197027.1|PREDICTED: nucleoporin p58/p45 isoform X1 [Lepisosteus oculatus] 6e-36; | ||||||
| NvERTx.4.88137 | R-7 | 0.9998327724 | E-5 | 0.999999999999 | Q8R332 | Nucleoporin p58/p45 {ECO:0000305} | gi|999984826|gb|KXJ23967.1|Nucleoporin p58/p45 [Exaiptasia pallida], 1e-90 |
| other nr_hits | gi|156365715|ref|XP_001626789.1|predicted protein [Nematostella vectensis] 4e-72; gi|919083271|ref|XP_013378988.1|PREDICTED: nucleoporin p58/p45-like isoform X1 [Lingula anatina] 1e-57; gi|919083274|ref|XP_013378989.1|PREDICTED: nucleoporin p58/p45-like isoform X2 [Lingula anatina] 1e-57; gi|1139831278|ref|XP_019929192.1|PREDICTED: probable nucleoporin Nup58 [Crassostrea gigas] 4e-55; gi|537241758|gb|ERE87297.1|nucleoporin [Cricetulus griseus] 6e-54; gi|537241761|gb|ERE87300.1|nucleoporin [Cricetulus griseus] 6e-54; gi|975118868|ref|XP_015274854.1|PREDICTED: nucleoporin p58/p45 isoform X2 [Gekko japonicus] 1e-53; gi|585712152|ref|XP_006824869.1|PREDICTED: nucleoporin p58/p45-like [Saccoglossus kowalevskii] 3e-53; gi|637338892|ref|XP_008116080.1|PREDICTED: nucleoporin p58/p45 isoform X2 [Anolis carolinensis] 3e-53; | ||||||
| NvERTx.4.112216 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q91876 | DNA replication licensing factor mcm7-A | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 6e-271 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 3e-243; gi|942201239|ref|XP_014352564.1|PREDICTED: DNA replication licensing factor MCM7 isoform X2 [Latimeria chalumnae] 3e-212; gi|557019447|ref|XP_006010077.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Latimeria chalumnae] 3e-212; gi|1072269289|ref|XP_018420309.1|PREDICTED: DNA replication licensing factor MCM7 [Nanorana parkeri] 2e-210; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 2e-209; gi|148232250|ref|NP_001081466.1|DNA replication licensing factor mcm7-A [Xenopus laevis] 3e-209; gi|109940098|sp|MCM7A_XENLA|Q91876.2|RecName: Full=DNA replication licensing factor mcm7-A; AltName: Full=CDC47 homolog A; AltName: Full=CDC47p; AltName: Full=Minichromosome maintenance protein 7-A; Short=xMCM7-A; AltName: Full=p90 >AAH72932.1 LOC397852 protein [Xenopus laevis] 8e-209; gi|82241532|sp|MCM7B_XENLA|Q7ZXB1.1|RecName: Full=DNA replication licensing factor mcm7-B; AltName: Full=CDC47 homolog B; AltName: Full=CDC47-2p; AltName: Full=Minichromosome maintenance protein 7-B; Short=xMCM7-B >AAH45072.1 Mcm7-prov protein [Xenopus laevis] 1e-208; gi|47498066|ref|NP_998877.1|DNA replication licensing factor mcm7 [Xenopus tropicalis] 1e-208; | ||||||
| NvERTx.4.112225 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.112217 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q3ZBH9 | DNA replication licensing factor MCM7 | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 6e-41 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 5e-34; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 2e-25; gi|1005438407|ref|XP_015756606.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Acropora digitifera] 2e-21; gi|195996431|ref|XP_002108084.1|hypothetical protein TRIADDRAFT_19118 [Trichoplax adhaerens] 2e-19; gi|731514002|ref|XP_010600113.1|PREDICTED: DNA replication licensing factor MCM7 isoform X2 [Loxodonta africana] 4e-19; gi|731514004|ref|XP_010600114.1|PREDICTED: DNA replication licensing factor MCM7 isoform X3 [Loxodonta africana] 4e-19; gi|344307766|ref|XP_003422550.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Loxodonta africana] 4e-19; gi|941787520|ref|XP_014325370.1|PREDICTED: DNA replication licensing factor MCM7 [Xiphophorus maculatus] 5e-19; gi|340380979|ref|XP_003388999.1|PREDICTED: DNA replication licensing factor mcm7-like [Amphimedon queenslandica] 6e-19; | ||||||
| NvERTx.4.112218 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q3ZBH9 | DNA replication licensing factor MCM7 | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 1e-118 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 1e-92; gi|291239877|ref|XP_002739848.1|PREDICTED: DNA replication licensing factor mcm7-like [Saccoglossus kowalevskii] 9e-74; gi|632988224|ref|XP_007882990.1|PREDICTED: DNA replication licensing factor MCM7 [Callorhinchus milii] 1e-73; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 2e-73; gi|961940028|ref|XP_014913365.1|PREDICTED: DNA replication licensing factor MCM7 isoform X2 [Poecilia latipinna] 6e-73; gi|961940032|ref|XP_014913366.1|PREDICTED: DNA replication licensing factor MCM7 isoform X3 [Poecilia latipinna] 6e-73; gi|961940024|ref|XP_014913364.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia latipinna] 6e-73; gi|961944409|ref|XP_014867403.1|PREDICTED: DNA replication licensing factor MCM7 [Poecilia mexicana] 2e-72; gi|658871613|ref|XP_008418745.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia reticulata] 2e-72; | ||||||
| NvERTx.4.112219 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q6NX31 | DNA replication licensing factor mcm7 | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 0e+00; gi|557019447|ref|XP_006010077.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Latimeria chalumnae] 1e-293; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 2e-292; gi|47498066|ref|NP_998877.1|DNA replication licensing factor mcm7 [Xenopus tropicalis] 2e-288; gi|82241532|sp|MCM7B_XENLA|Q7ZXB1.1|RecName: Full=DNA replication licensing factor mcm7-B; AltName: Full=CDC47 homolog B; AltName: Full=CDC47-2p; AltName: Full=Minichromosome maintenance protein 7-B; Short=xMCM7-B >AAH45072.1 Mcm7-prov protein [Xenopus laevis] 2e-288; gi|148232250|ref|NP_001081466.1|DNA replication licensing factor mcm7-A [Xenopus laevis] 6e-288; gi|961940024|ref|XP_014913364.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia latipinna] 1e-287; gi|961944409|ref|XP_014867403.1|PREDICTED: DNA replication licensing factor MCM7 [Poecilia mexicana] 1e-287; gi|658871613|ref|XP_008418745.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia reticulata] 2e-287; | ||||||
| NvERTx.4.112220 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q3ZBH9 | DNA replication licensing factor MCM7 | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 6e-41 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 5e-34; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 2e-25; gi|1005438407|ref|XP_015756606.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Acropora digitifera] 2e-21; gi|195996431|ref|XP_002108084.1|hypothetical protein TRIADDRAFT_19118 [Trichoplax adhaerens] 2e-19; gi|731514002|ref|XP_010600113.1|PREDICTED: DNA replication licensing factor MCM7 isoform X2 [Loxodonta africana] 4e-19; gi|731514004|ref|XP_010600114.1|PREDICTED: DNA replication licensing factor MCM7 isoform X3 [Loxodonta africana] 4e-19; gi|344307766|ref|XP_003422550.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Loxodonta africana] 4e-19; gi|941787520|ref|XP_014325370.1|PREDICTED: DNA replication licensing factor MCM7 [Xiphophorus maculatus] 5e-19; gi|340380979|ref|XP_003388999.1|PREDICTED: DNA replication licensing factor mcm7-like [Amphimedon queenslandica] 6e-19; | ||||||
| NvERTx.4.112221 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q61881 | DNA replication licensing factor MCM7 | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 3e-54 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 2e-45; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 7e-35; gi|1049243723|ref|XP_017560902.1|PREDICTED: DNA replication licensing factor MCM7 [Pygocentrus nattereri] 6e-29; gi|941787520|ref|XP_014325370.1|PREDICTED: DNA replication licensing factor MCM7 [Xiphophorus maculatus] 1e-28; gi|1067117321|ref|XP_018014775.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hyalella azteca] 2e-28; gi|1025466690|ref|XP_016535552.1|PREDICTED: LOW QUALITY PROTEIN: DNA replication licensing factor MCM7 [Poecilia formosa] 2e-28; gi|961940024|ref|XP_014913364.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia latipinna] 2e-28; gi|961944409|ref|XP_014867403.1|PREDICTED: DNA replication licensing factor MCM7 [Poecilia mexicana] 2e-28; gi|942201239|ref|XP_014352564.1|PREDICTED: DNA replication licensing factor MCM7 isoform X2 [Latimeria chalumnae] 3e-28; | ||||||
| NvERTx.4.112222 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q91876 | DNA replication licensing factor mcm7-A | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 6e-271 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 3e-243; gi|942201239|ref|XP_014352564.1|PREDICTED: DNA replication licensing factor MCM7 isoform X2 [Latimeria chalumnae] 3e-212; gi|557019447|ref|XP_006010077.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Latimeria chalumnae] 3e-212; gi|1072269289|ref|XP_018420309.1|PREDICTED: DNA replication licensing factor MCM7 [Nanorana parkeri] 2e-210; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 2e-209; gi|148232250|ref|NP_001081466.1|DNA replication licensing factor mcm7-A [Xenopus laevis] 3e-209; gi|109940098|sp|MCM7A_XENLA|Q91876.2|RecName: Full=DNA replication licensing factor mcm7-A; AltName: Full=CDC47 homolog A; AltName: Full=CDC47p; AltName: Full=Minichromosome maintenance protein 7-A; Short=xMCM7-A; AltName: Full=p90 >AAH72932.1 LOC397852 protein [Xenopus laevis] 8e-209; gi|82241532|sp|MCM7B_XENLA|Q7ZXB1.1|RecName: Full=DNA replication licensing factor mcm7-B; AltName: Full=CDC47 homolog B; AltName: Full=CDC47-2p; AltName: Full=Minichromosome maintenance protein 7-B; Short=xMCM7-B >AAH45072.1 Mcm7-prov protein [Xenopus laevis] 1e-208; gi|47498066|ref|NP_998877.1|DNA replication licensing factor mcm7 [Xenopus tropicalis] 1e-208; | ||||||
| NvERTx.4.112223 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q6NX31 | DNA replication licensing factor mcm7 | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 0e+00 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 0e+00; gi|557019447|ref|XP_006010077.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Latimeria chalumnae] 1e-293; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 2e-292; gi|47498066|ref|NP_998877.1|DNA replication licensing factor mcm7 [Xenopus tropicalis] 2e-288; gi|82241532|sp|MCM7B_XENLA|Q7ZXB1.1|RecName: Full=DNA replication licensing factor mcm7-B; AltName: Full=CDC47 homolog B; AltName: Full=CDC47-2p; AltName: Full=Minichromosome maintenance protein 7-B; Short=xMCM7-B >AAH45072.1 Mcm7-prov protein [Xenopus laevis] 2e-288; gi|148232250|ref|NP_001081466.1|DNA replication licensing factor mcm7-A [Xenopus laevis] 6e-288; gi|961940024|ref|XP_014913364.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia latipinna] 1e-287; gi|961944409|ref|XP_014867403.1|PREDICTED: DNA replication licensing factor MCM7 [Poecilia mexicana] 1e-287; gi|658871613|ref|XP_008418745.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia reticulata] 2e-287; | ||||||
| NvERTx.4.112224 | R-5 | 0.999003249869 | E-5 | 0.999999999999 | Q61881 | DNA replication licensing factor MCM7 | gi|156375279|ref|XP_001630009.1|predicted protein [Nematostella vectensis], 3e-54 |
| other nr_hits | gi|999985835|gb|KXJ24976.1|DNA replication licensing factor MCM7 [Exaiptasia pallida] 2e-45; gi|828216528|ref|XP_012560346.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hydra vulgaris] 7e-35; gi|1049243723|ref|XP_017560902.1|PREDICTED: DNA replication licensing factor MCM7 [Pygocentrus nattereri] 6e-29; gi|941787520|ref|XP_014325370.1|PREDICTED: DNA replication licensing factor MCM7 [Xiphophorus maculatus] 1e-28; gi|1067117321|ref|XP_018014775.1|PREDICTED: DNA replication licensing factor mcm7-B-like [Hyalella azteca] 2e-28; gi|1025466690|ref|XP_016535552.1|PREDICTED: LOW QUALITY PROTEIN: DNA replication licensing factor MCM7 [Poecilia formosa] 2e-28; gi|961940024|ref|XP_014913364.1|PREDICTED: DNA replication licensing factor MCM7 isoform X1 [Poecilia latipinna] 2e-28; gi|961944409|ref|XP_014867403.1|PREDICTED: DNA replication licensing factor MCM7 [Poecilia mexicana] 2e-28; gi|942201239|ref|XP_014352564.1|PREDICTED: DNA replication licensing factor MCM7 isoform X2 [Latimeria chalumnae] 3e-28; | ||||||
| NvERTx.4.95813 | R-8 | 0.997076713216 | E-5 | 0.999999999999 | O95995 | Growth arrest-specific protein 8 | gi|156385510|ref|XP_001633673.1|predicted protein [Nematostella vectensis], 2e-222 |
| other nr_hits | gi|999971684|gb|KXJ10858.1|Growth arrest-specific protein 8 [Exaiptasia pallida] 3e-212; gi|1005476117|ref|XP_015774822.1|PREDICTED: growth arrest-specific protein 8-like [Acropora digitifera] 1e-211; gi|260834855|ref|XP_002612425.1|hypothetical protein BRAFLDRAFT_263599 [Branchiostoma floridae] 3e-199; gi|919016103|ref|XP_013392403.1|PREDICTED: growth arrest-specific protein 8-like [Lingula anatina] 7e-199; gi|1126192771|ref|XP_019629401.1|PREDICTED: growth arrest-specific protein 8-like [Branchiostoma belcheri] 1e-198; gi|291227693|ref|XP_002733817.1|PREDICTED: growth arrest-specific protein 8-like [Saccoglossus kowalevskii] 4e-195; gi|676456308|ref|XP_009054282.1|hypothetical protein LOTGIDRAFT_215253 [Lottia gigantea] 5e-194; gi|443713227|gb|ELU06192.1|hypothetical protein CAPTEDRAFT_162685 [Capitella teleta] 1e-192; gi|524879981|ref|XP_005096659.1|PREDICTED: growth arrest-specific protein 8-like isoform X1 [Aplysia californica] 5e-190; | ||||||
| NvERTx.4.95814 | R-8 | 0.997076713216 | E-5 | 0.999999999999 | O95995 | Growth arrest-specific protein 8 | gi|156385510|ref|XP_001633673.1|predicted protein [Nematostella vectensis], 2e-126 |
| other nr_hits | gi|1005476117|ref|XP_015774822.1|PREDICTED: growth arrest-specific protein 8-like [Acropora digitifera] 8e-123; gi|999971684|gb|KXJ10858.1|Growth arrest-specific protein 8 [Exaiptasia pallida] 4e-121; gi|1126192771|ref|XP_019629401.1|PREDICTED: growth arrest-specific protein 8-like [Branchiostoma belcheri] 3e-114; gi|260834855|ref|XP_002612425.1|hypothetical protein BRAFLDRAFT_263599 [Branchiostoma floridae] 3e-114; gi|919016103|ref|XP_013392403.1|PREDICTED: growth arrest-specific protein 8-like [Lingula anatina] 2e-113; gi|291227693|ref|XP_002733817.1|PREDICTED: growth arrest-specific protein 8-like [Saccoglossus kowalevskii] 2e-112; gi|676456308|ref|XP_009054282.1|hypothetical protein LOTGIDRAFT_215253 [Lottia gigantea] 2e-112; gi|871227196|ref|XP_012936950.1|PREDICTED: growth arrest-specific protein 8-like isoform X2 [Aplysia californica] 3e-110; gi|524879981|ref|XP_005096659.1|PREDICTED: growth arrest-specific protein 8-like isoform X1 [Aplysia californica] 3e-110; | ||||||
| NvERTx.4.199577 | R-5 | 0.999973166179 | E-5 | 0.999999999999 | P49454 | Centromere protein F | gi|999976248|gb|KXJ15422.1|Centromere protein F [Exaiptasia pallida], 2e-266 |
| other nr_hits | gi|762136508|ref|XP_011451531.1|PREDICTED: centromere protein F [Crassostrea gigas] 5e-163; gi|405966865|gb|EKC32097.1|Centromere protein F [Crassostrea gigas] 5e-163; gi|390341494|ref|XP_796801.2|PREDICTED: centromere protein F [Strongylocentrotus purpuratus] 2e-159; gi|919067218|ref|XP_013415148.1|PREDICTED: centromere protein F-like [Lingula anatina] 1e-156; gi|688600508|ref|XP_009292627.1|PREDICTED: nesprin-1 isoform X3 [Danio rerio] 2e-137; gi|528510293|ref|XP_685984.6|PREDICTED: nesprin-1 isoform X1 [Danio rerio] 2e-137; gi|528510301|ref|XP_005159741.1|PREDICTED: nesprin-1 isoform X4 [Danio rerio] 2e-137; gi|154413468|ref|XP_001579764.1|viral A-type inclusion protein [Trichomonas vaginalis G3] 1e-136; gi|528510297|ref|XP_005159739.1|PREDICTED: nesprin-1 isoform X2 [Danio rerio] 1e-135; | ||||||
| NvERTx.4.140911 | R-7 | 0.962583260401 | E-5 | 0.999999999999 | Q4U0S5 | RNA polymerase II-associated factor 1 homolog | gi|156389561|ref|XP_001635059.1|predicted protein [Nematostella vectensis], 2e-165 |
| other nr_hits | gi|999978568|gb|KXJ17742.1|RNA polymerase II-associated factor 1-like [Exaiptasia pallida] 3e-151; gi|1126186579|ref|XP_019626009.1|PREDICTED: LOW QUALITY PROTEIN: RNA polymerase II-associated factor 1 homolog [Branchiostoma belcheri] 1e-110; gi|1005480248|ref|XP_015776857.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Acropora digitifera] 9e-110; gi|769864576|ref|XP_011644176.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Pogonomyrmex barbatus] 9e-110; gi|749733415|ref|XP_011145835.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Harpegnathos saltator] 9e-110; gi|780711212|ref|XP_011704570.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Wasmannia auropunctata] 1e-109; gi|1070183350|ref|XP_018365534.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Trachymyrmex cornetzi] 2e-109; gi|1070183354|ref|XP_018365535.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X3 [Trachymyrmex cornetzi] 2e-109; gi|1069714715|ref|XP_018314327.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X1 [Trachymyrmex zeteki] 2e-109; | ||||||
| NvERTx.4.140912 | R-7 | 0.962583260401 | E-5 | 0.999999999999 | Q5RAX0 | RNA polymerase II-associated factor 1 homolog | gi|156389561|ref|XP_001635059.1|predicted protein [Nematostella vectensis], 3e-187 |
| other nr_hits | gi|999978568|gb|KXJ17742.1|RNA polymerase II-associated factor 1-like [Exaiptasia pallida] 2e-174; gi|1005480248|ref|XP_015776857.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Acropora digitifera] 4e-131; gi|584013672|ref|XP_006801626.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X3 [Neolamprologus brichardi] 8e-126; gi|548404604|ref|XP_005739415.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Pundamilia nyererei] 8e-126; gi|499014365|ref|XP_004558212.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Maylandia zebra] 8e-126; gi|974052222|ref|XP_015239209.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Cyprinodon variegatus] 1e-125; gi|961911539|ref|XP_014858778.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Poecilia mexicana] 1e-125; gi|658880477|ref|XP_008423175.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Poecilia reticulata] 1e-125; gi|617407724|ref|XP_007554421.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Poecilia formosa] 1e-125; | ||||||
| NvERTx.4.140913 | R-7 | 0.962583260401 | E-5 | 0.999999999999 | Q5RAX0 | RNA polymerase II-associated factor 1 homolog | gi|156389561|ref|XP_001635059.1|predicted protein [Nematostella vectensis], 3e-187 |
| other nr_hits | gi|999978568|gb|KXJ17742.1|RNA polymerase II-associated factor 1-like [Exaiptasia pallida] 2e-174; gi|1005480248|ref|XP_015776857.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Acropora digitifera] 4e-131; gi|584013672|ref|XP_006801626.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X3 [Neolamprologus brichardi] 8e-126; gi|548404604|ref|XP_005739415.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Pundamilia nyererei] 8e-126; gi|499014365|ref|XP_004558212.1|PREDICTED: RNA polymerase II-associated factor 1 homolog [Maylandia zebra] 8e-126; gi|974052222|ref|XP_015239209.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Cyprinodon variegatus] 1e-125; gi|961911539|ref|XP_014858778.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Poecilia mexicana] 1e-125; gi|658880477|ref|XP_008423175.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Poecilia reticulata] 1e-125; gi|617407724|ref|XP_007554421.1|PREDICTED: RNA polymerase II-associated factor 1 homolog isoform X2 [Poecilia formosa] 1e-125; | ||||||
| NvERTx.4.29630 | R-7 | 0.993098065454 | E-5 | 0.999999999999 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
| NvERTx.4.29631 | R-7 | 0.993098065454 | E-5 | 0.999999999999 | No_Uniprotmatch | No_Description | None |
| other nr_hits | NA | ||||||
